The R package visStatistics allows for rapid visualisation and statistical analysis of raw data. It automatically selects and visualises the most appropriate statistical hypothesis test between two vectors of class integer, numeric or factor.
While numerous R packages provide statistical testing functionality, few are designed with pedagogical accessibility as a primary concern. visStatistics addresses this gap by automating the test selection process and presenting results using annotated, publication-ready visualisations. This helps the user to focus on interpretation rather than technical execution.
The automated workflow is particularly suited for browser-based interfaces that rely on server-side R applications connected to secure databases, where users have no direct access, or for quick data visualisation, e.g. in statistical consulting projects or educational settings.
Getting Started
The function visstat() accepts input in two ways:
# Standardised form (recommended):
visstat(x, y)
# Backward-compatible form:
visstat(dataframe, "namey", "namex")In the standardised form, x and y must be vectors of class "numeric", "integer", or "factor".
In the backward-compatible form, "namex" and "namey" must be character strings naming columns in a data.frame named dataframe. These column must be of class "numeric", "integer", or "factor". This is equivalent to writing:
To simplify the notation, throughout the remainder, data of class numeric or integer are both referred to by their common mode numeric, while data of class factor are referred to as categorical.
The interpretation of x and y depends on their classes:
If one is numeric and the other is a factor, the numeric must be passed as response
yand the factor as predictorx. This supports tests for central tendencies.If both are numeric, a simple linear regression model is fitted with
yas the response andxas the predictor.If both are factors, a test of association is performed (Chi-squared or Fisher’s exact). The test is symmetric, but the plot layout depends on which variable is supplied as
x.
visstat() selects the appropriate statistical test and generates visualisations accompanied by the main test statistics.
Examples
Numerical response and categorical predictor
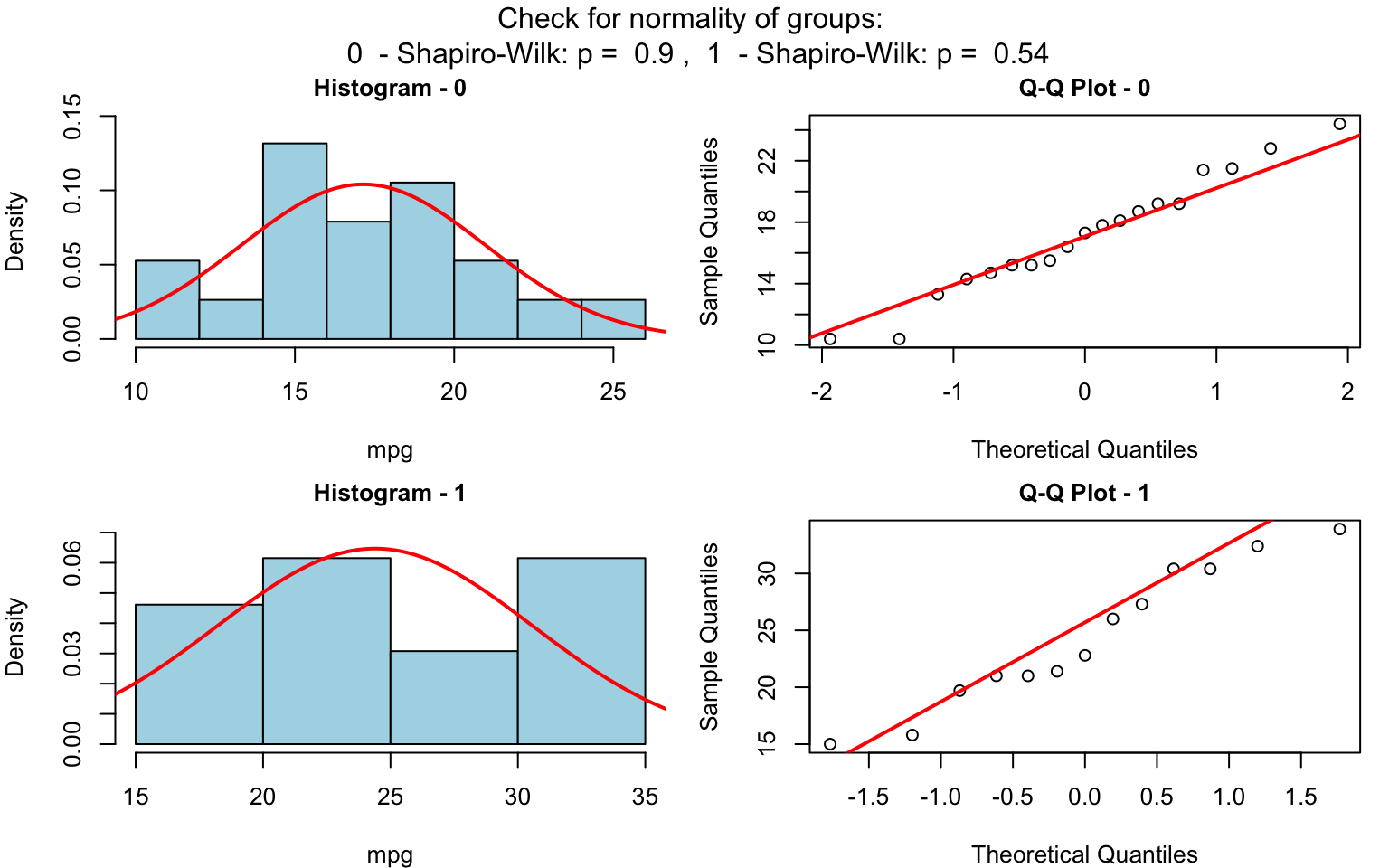
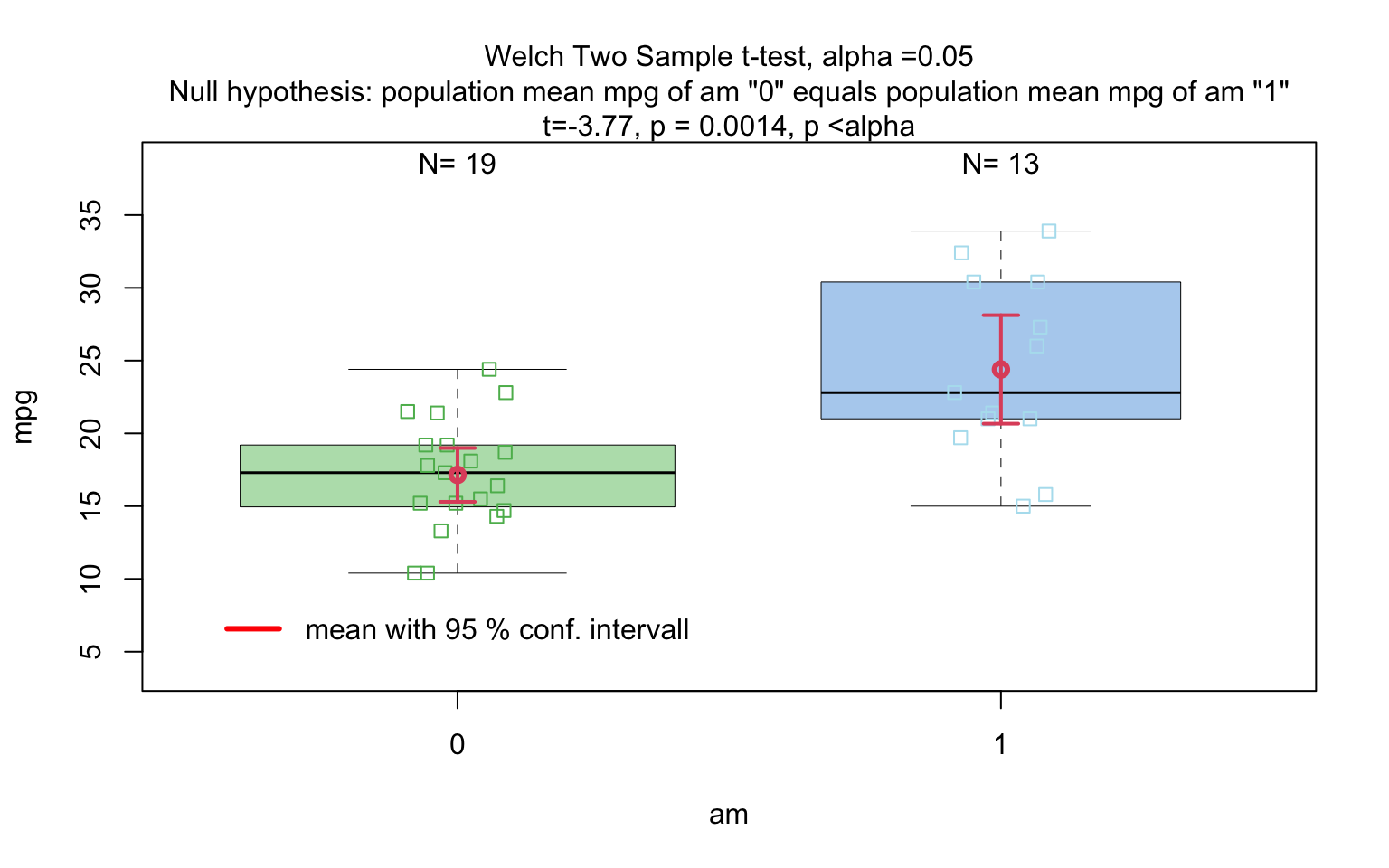
When the response is numerical and the predictor is categorical, test of central tendencies are selected.
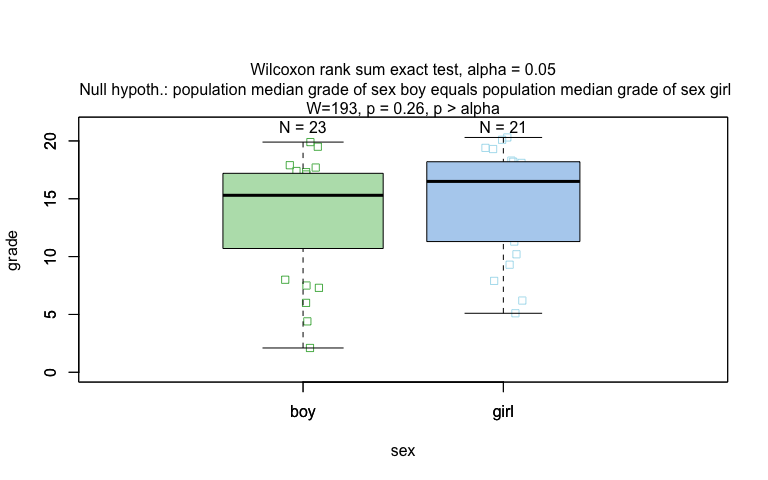
Wilcoxon rank sum test
grades_gender <- data.frame(
sex = factor(rep(c("girl", "boy"), times = c(21, 23))),
grade = c(
19.3, 18.1, 15.2, 18.3, 7.9, 6.2, 19.4, 20.3, 9.3, 11.3,
18.2, 17.5, 10.2, 20.1, 13.3, 17.2, 15.1, 16.2, 17.0, 16.5, 5.1,
15.3, 17.1, 14.8, 15.4, 14.4, 7.5, 15.5, 6.0, 17.4, 7.3, 14.3,
13.5, 8.0, 19.5, 13.4, 17.9, 17.7, 16.4, 15.6, 17.3, 19.9, 4.4, 2.1
)
)
wilcoxon_statistics <- visstat(grades_gender$sex, grades_gender$grade)
Numerical response and numerical predictor: Linear Regression
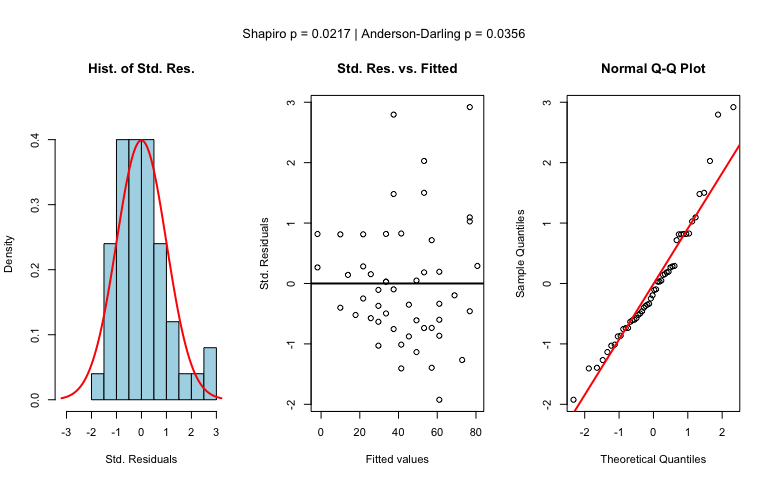
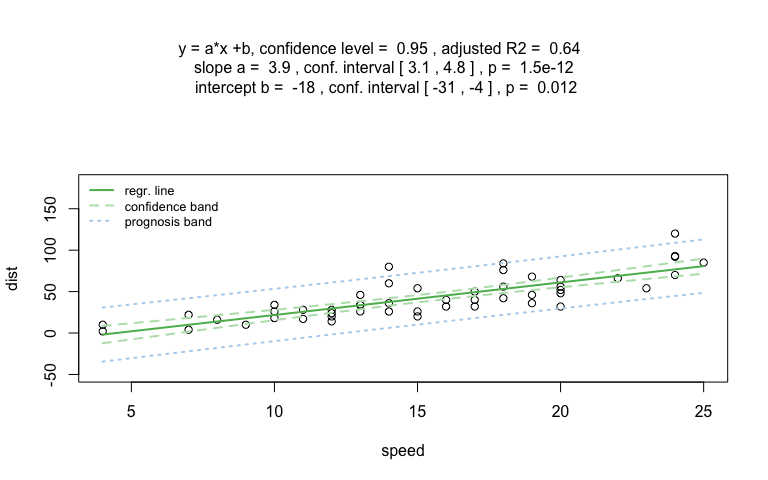
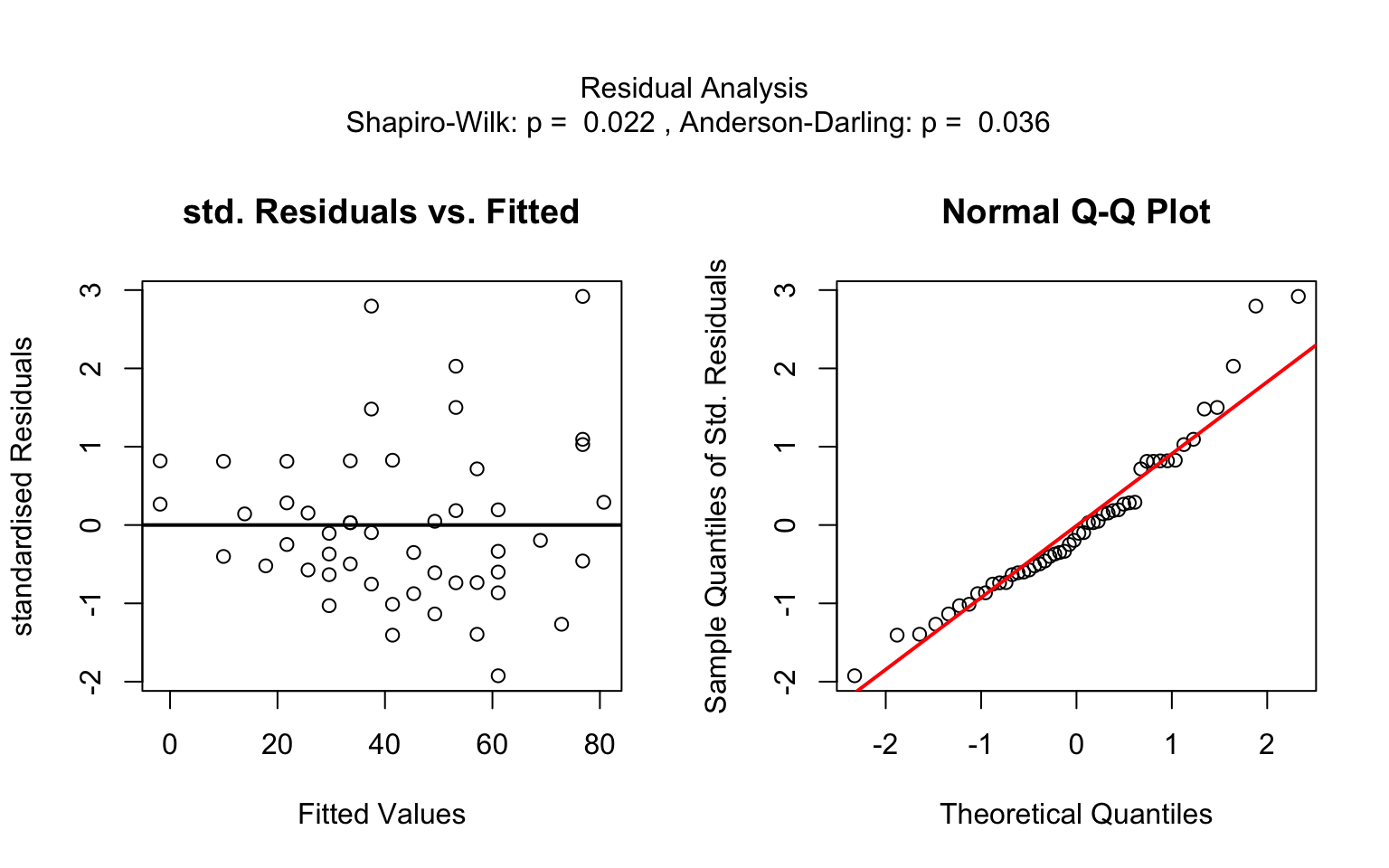
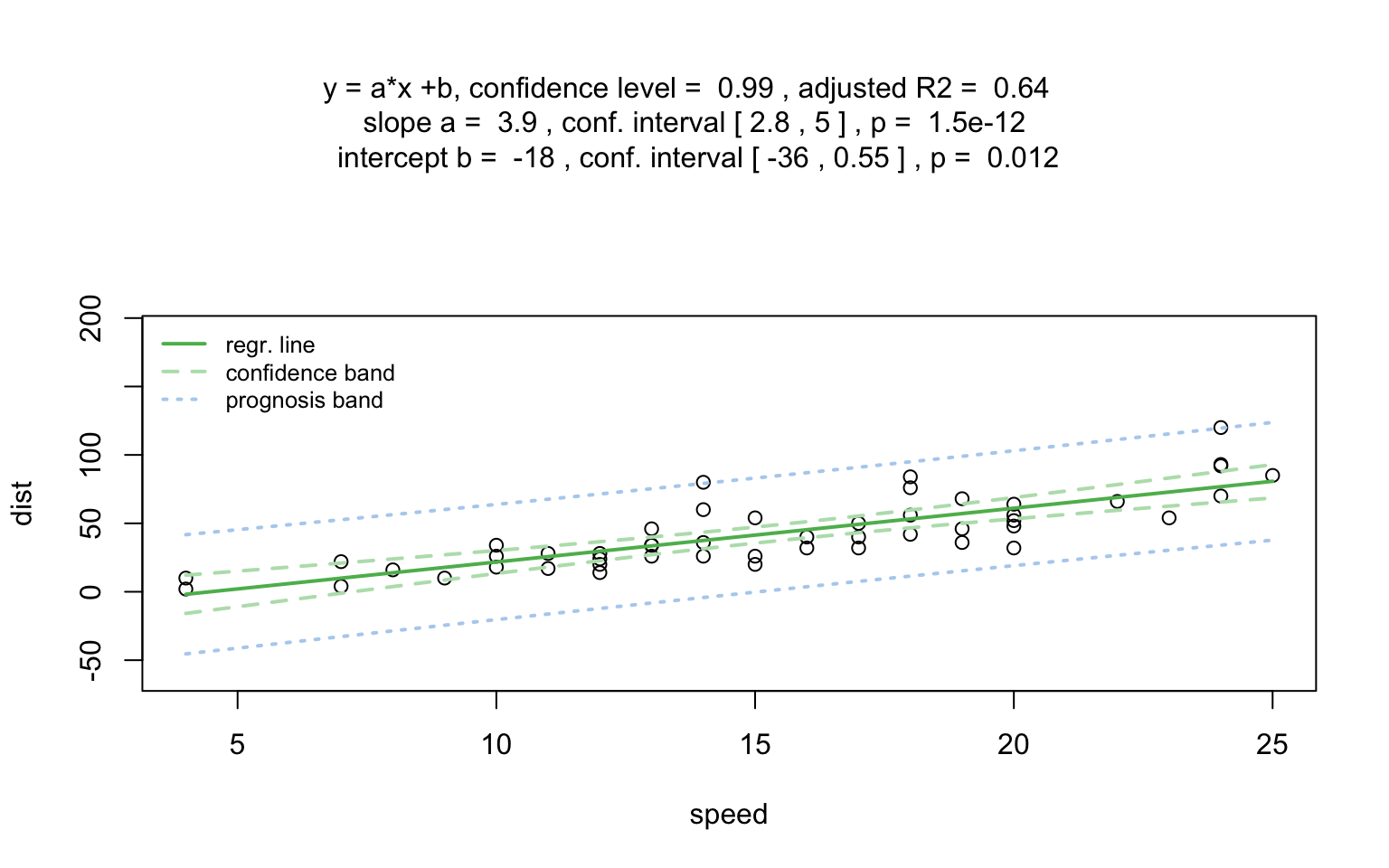
Increasing the confidence level conf.level from the default 0.95 to 0.99 leads two wider confidence and prediction bands:
Both variables categorical
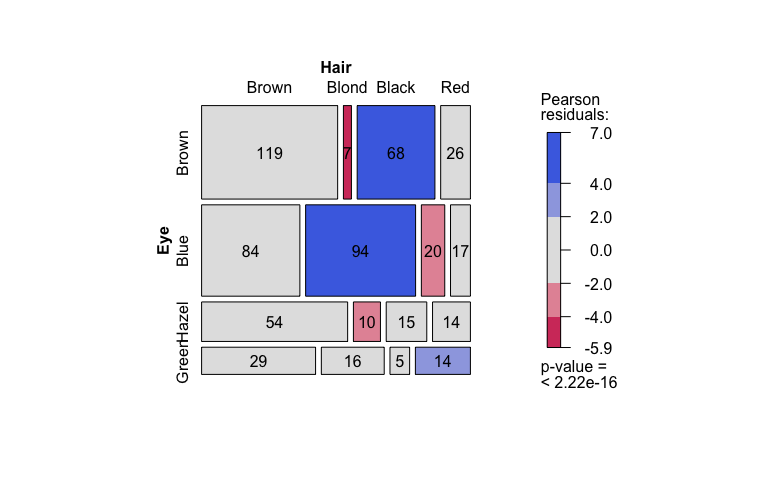
Pearson’s Chi-squared test
Count data sets are often presented as multidimensional arrays, so - called contingency tables, whereas visstat() requires a data.frame with a column structure. Arrays can be transformed to this column wise structure with the helper function counts_to_cases():
hair_eye_color_df <- counts_to_cases(as.data.frame(HairEyeColor))
visstat(hair_eye_color_df$Eye, hair_eye_color_df$Hair)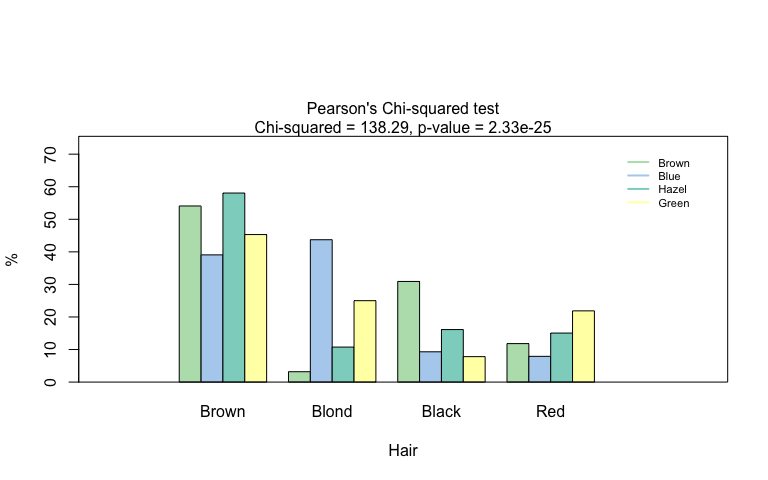
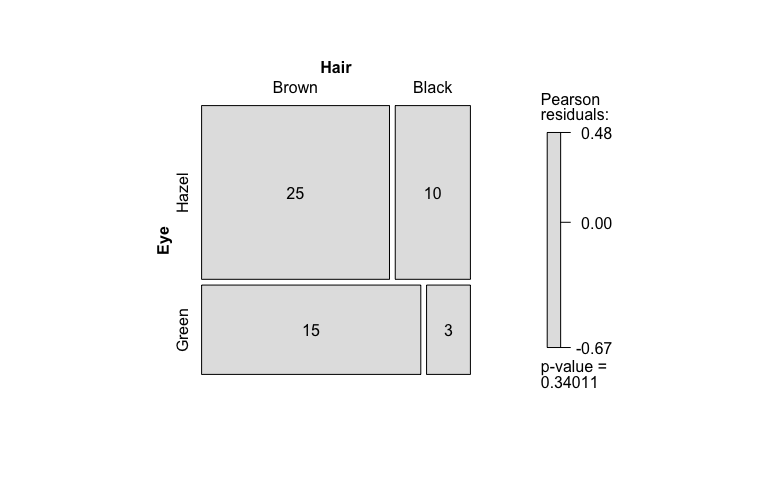
Fisher’s exact test
hair_eye_color_male <- HairEyeColor[, , 1]
# Slice out a 2 by 2 contingency table
black_brown_hazel_green_male <- hair_eye_color_male[1:2, 3:4]
# Transform to data frame
black_brown_hazel_green_male <- counts_to_cases(as.data.frame(black_brown_hazel_green_male))
# Fisher test
fisher_stats <- visstat(black_brown_hazel_green_male$Eye,black_brown_hazel_green_male$Hair)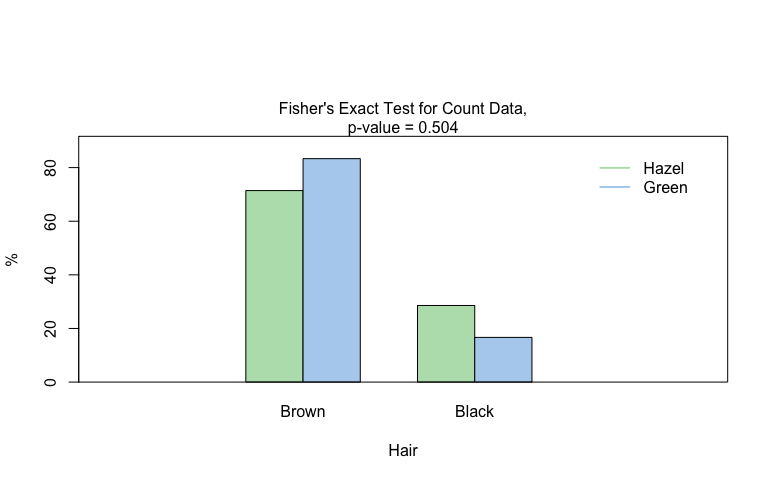
Saving the graphical output
All generated graphics can be saved in any file format supported by Cairo(), including “png”, “jpeg”, “pdf”, “svg”, “ps”, and “tiff” in the user specified plotDirectory.
If the optional argument plotName is not given, the naming of the output follows the pattern "testname_namey_namex.", where "testname" specifies the selected test or visualisation and "namey" and "namex" are character strings naming the selected data vectors y and x, respectively. The suffix corresponding to the chosen graphicsoutput (e.g., "pdf", "png") is then concatenated to form the complete output file name.
In the following example, we store the graphics in png format in the plotDirectory tempdir() with the default naming convention:
#Graphical output written to plotDirectory: In this example
# a bar chart to visualise the Chi-squared test and mosaic plot showing
# Pearson's residuals named
#chi_squared_or_fisher_Hair_Eye.png and mosaic_complete_Hair_Eye.png resp.
save_fisher <- visstat(black_brown_hazel_green_male, "Hair", "Eye",
graphicsoutput = "png", plotDirectory = tempdir())The full file path of the generated graphics are stored as the attribute "plot_paths" on the returned object of class "visstat".
paths <- attr(save_fisher, "plot_paths")
print(paths)
#> [1] "/var/folders/5c/n85wqnh95l50qbp3s9l0rp_w0000gn/T//RtmpucUdua/chi_squared_or_fisher_Hair_Eye.png"
#> [2] "/var/folders/5c/n85wqnh95l50qbp3s9l0rp_w0000gn/T//RtmpucUdua/mosaic_complete_Hair_Eye.png"Remove the graphical output from plotDirectory:
Decision logic
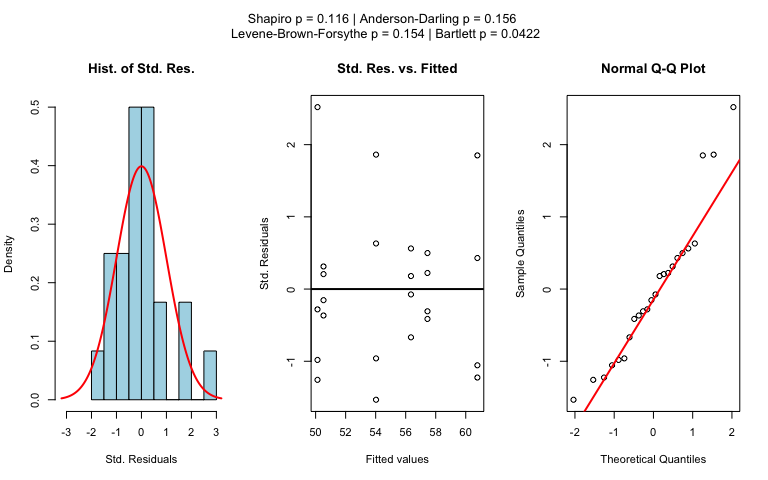
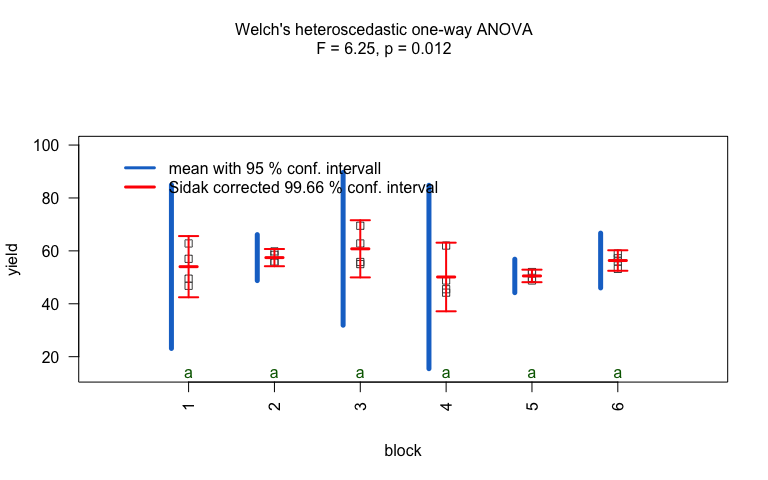
The choice of statistical tests depends on whether the data of the selected columns are numeric or categorical, the number of levels in the categorical variable, and the distribution of the data.
For a detailed description of the underlying decision logic see
A graphical summary of the decision logic used for numerical responses and categorial predictors is given in below figure:
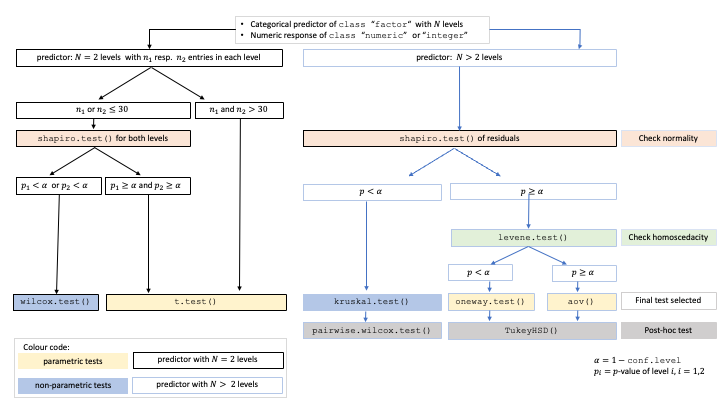
Decision tree used to select the appropriate statistical test for a categorical predictor and numeric response, based on the number of factor levels, normality, and homoscedasticity.
Implemented tests
Numerical response and categorical predictor
Main tests
t.test(), wilcox.test(), aov(), oneway.test(), kruskal.test()
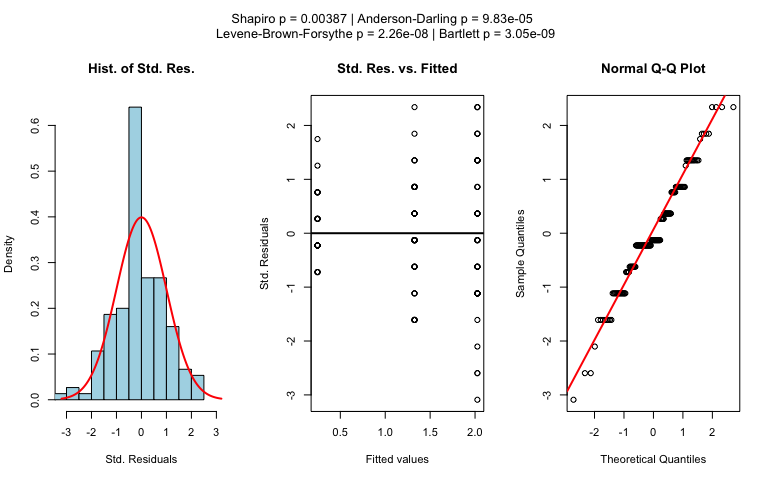
Normality assumption check
shapiro.test() and ad.test()(Gross and Ligges 2015)
Homoscedasticity assumption check
levene.test() and bartlett.test()
Post-hoc tests
-TukeyHSD() (used following aov()and oneway.test())
-pairwise.wilcox.test() (used following kruskal.test())
References
Gross, Juergen, and Uwe Ligges. 2015. Nortest: Tests for Normality. Manual. https://doi.org/10.32614/CRAN.package.nortest.