visstat() is a wrapper around visstat_core
that provides three alternative input styles: a formula interface, a
standardised vector interface, and a backward-compatible data frame interface.
visstat_core defines the decision logic for statistical
hypothesis testing and visualisation between two variables of class
"numeric", "integer", or "factor".
visstat(
x,
y,
...,
data = NULL,
conf.level = 0.95,
do_regression = TRUE,
numbers = TRUE,
minpercent = 0.05,
graphicsoutput = NULL,
plotName = NULL,
plotDirectory = getwd()
)Arguments
- x
For the formula interface: a formula of the form
y ~ x, whereyis the response variable andxis the predictor or grouping variable (requiresdataargument). For the standardised form: a vector of class"numeric","integer", or"factor"representing the predictor or grouping variable. For the backward-compatible form: adata.framecontaining the relevant columns.- y
For the formula interface: not used (variables are extracted from the formula). For the standardised form: a vector of class
"numeric","integer", or"factor"representing the response variable. For the backward-compatible form: acharacterstring specifying the name of the response variable column inx.- ...
For the backward-compatible form only: a
characterstring specifying the name of the predictor or grouping variable column inx. Ignored for formula and standardised input styles.- data
A
data.framecontaining the variables specified in the formula. Required when using the formula interface. Ignored for other input styles.- conf.level
Confidence level for statistical inference; default is
0.95.- do_regression
Logical. If
TRUE(default), performs simple linear regression analysis with confidence and prediction bands when both variables are numeric. IfFALSE, performs correlation analysis with trend line only (no regression interpretation).- numbers
Logical. Whether to annotate plots with numeric values.
- minpercent
Number between 0 and 1 indicating minimal fraction of total count data of a category to be displayed in mosaic count plots.
- graphicsoutput
Saves plot(s) of type
"png","jpg","tiff"or"bmp"in directory specified inplotDirectory. IfNULL, no plots are saved.- plotName
Graphical output is stored following the naming convention
"plotName.graphicsoutput"inplotDirectory. Without specifying this parameter,plotNameis automatically generated following the convention"statisticalTestName_varsample_varfactor".- plotDirectory
Specifies directory where generated plots are stored. Default is current working directory.
Value
An object of class "visstat" containing the results of
the automatically selected statistical test. The specific contents depend on
which test was performed. Additionally, the returned object includes two
attributes:
plot_paths: Character vector of file paths where plots were saved (ifgraphicsoutputwas specified)captured_plots: List of captured plot objects for programmatic access
In case of insufficient data, returns a list with an error element and
basic input summary information.
Details
This wrapper supports three input formats:
(1) Formula interface: visstat(y ~ x, data = df), where the formula
specifies the response (y) and predictor (x) variables, and
data is a data frame containing these variables.
(2) Standardised form: visstat(x, y), where both x and y
are vectors of class "numeric", "integer", or "factor".
Here x is the predictor or grouping variable and y is the
response variable.
(3) Backward-compatible form: visstat(dataframe, "name_of_y", "name_of_x"),
where the first character string refers to the response variable and the
second to the predictor or grouping variable. Both must be column names in
dataframe.
The interpretation of x and y depends on the variable classes.
Throughout, data of class numeric or integer are referred to
as numeric, while data of class factor are referred to as categorical:
If one variable is numeric and the other a factor, the numeric vector is the
response (y) and the factor is the grouping variable (x). This
supports tests of central tendencies (e.g., t-test, Welch's ANOVA, Wilcoxon,
Kruskal-Wallis).
If both variables are numeric, a linear model is fitted with y as the
response and x as the predictor.
If both variables are factors, an association test (Chi-squared or Fisher's
exact) is used. The test result is invariant to variable order, but
visualisations (e.g., axis layout, bar orientation) depend on the roles of
x and y.
This wrapper standardises the input and calls visstat_core,
which selects and executes the appropriate test with visual output and
assumption diagnostics.
Note
For best visualization, ensure the RStudio Plots pane is adequately
sized. If you get "figure margins too large" errors, try expanding the Plots
pane in RStudio, using dev.new(width=10, height=6) for a larger plot
window, or reducing the cex parameter.
See also
visstat_core defining the decision logic, the
package's vignette vignette("visStatistics") explaining the decision
logic accompanied by illustrative examples, and the accompanying webpage
https://shhschilling.github.io/visStatistics/.
Examples
# Formula interface
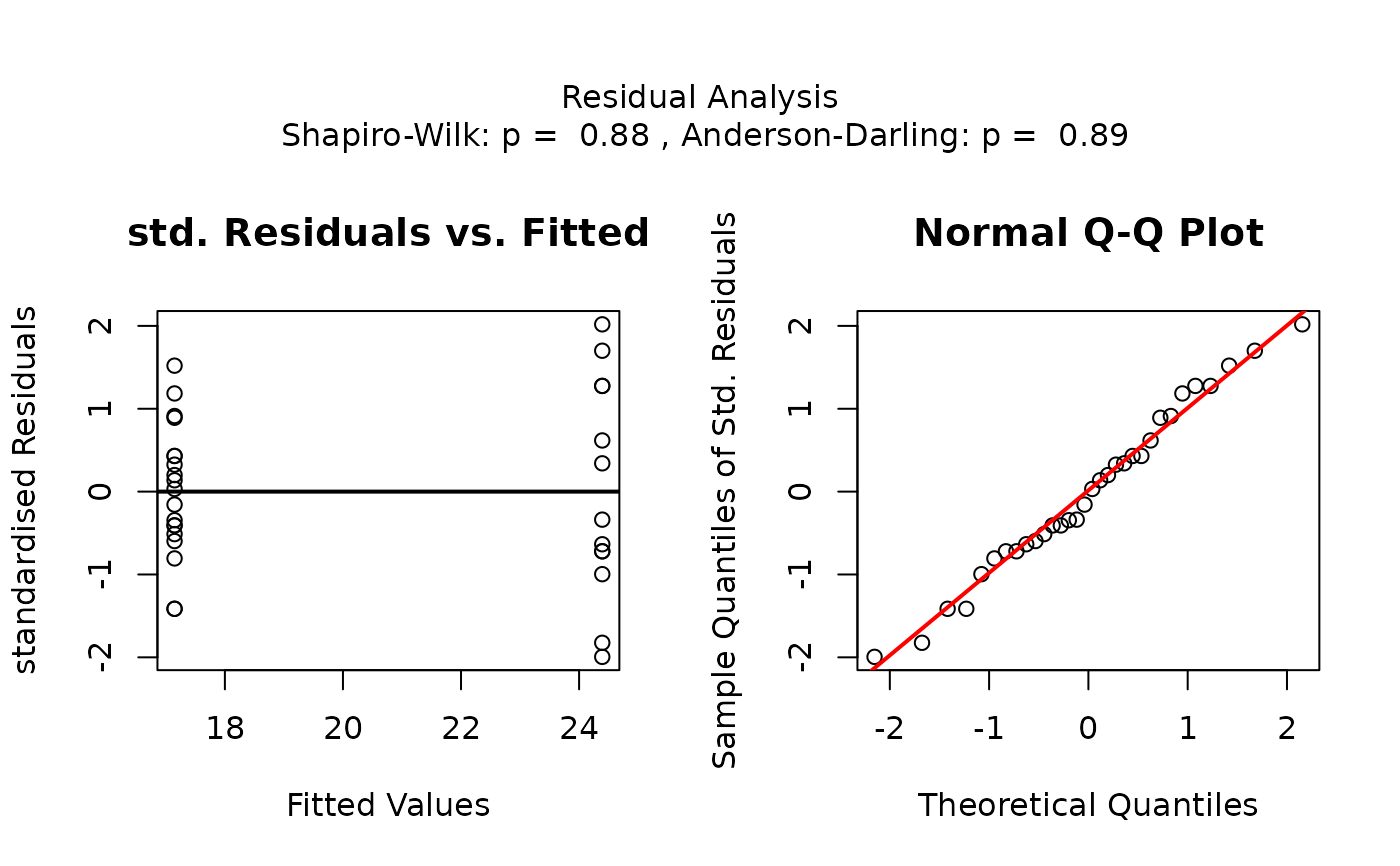
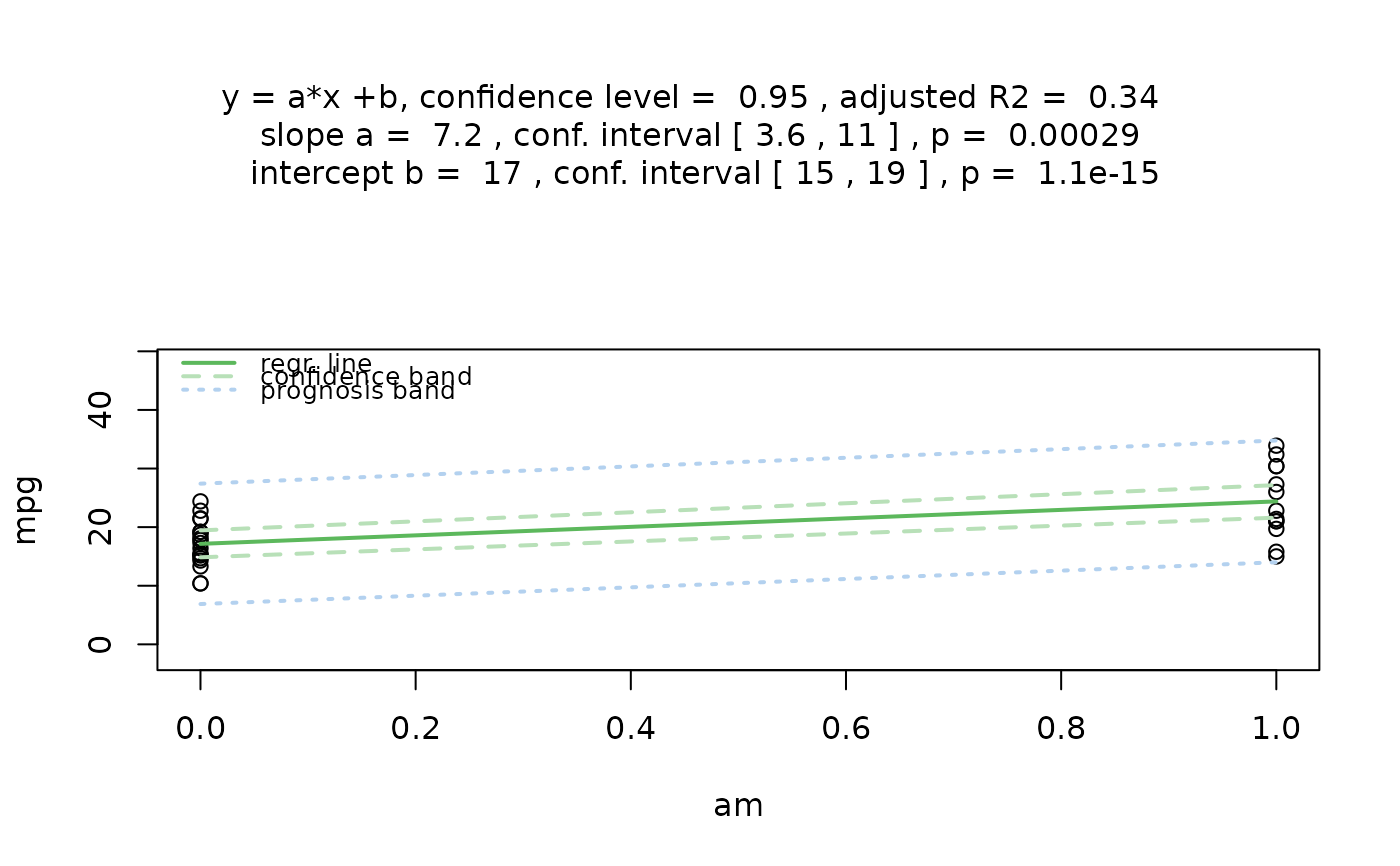
mtcars$am <- as.factor(mtcars$am)
visstat(mpg ~ am, data = mtcars)
 # Standardised usage
visstat(mtcars$am, mtcars$mpg)
# Standardised usage
visstat(mtcars$am, mtcars$mpg)

 # Backward-compatible usage (same result as above)
visstat(mtcars, "mpg", "am")
# Backward-compatible usage (same result as above)
visstat(mtcars, "mpg", "am")

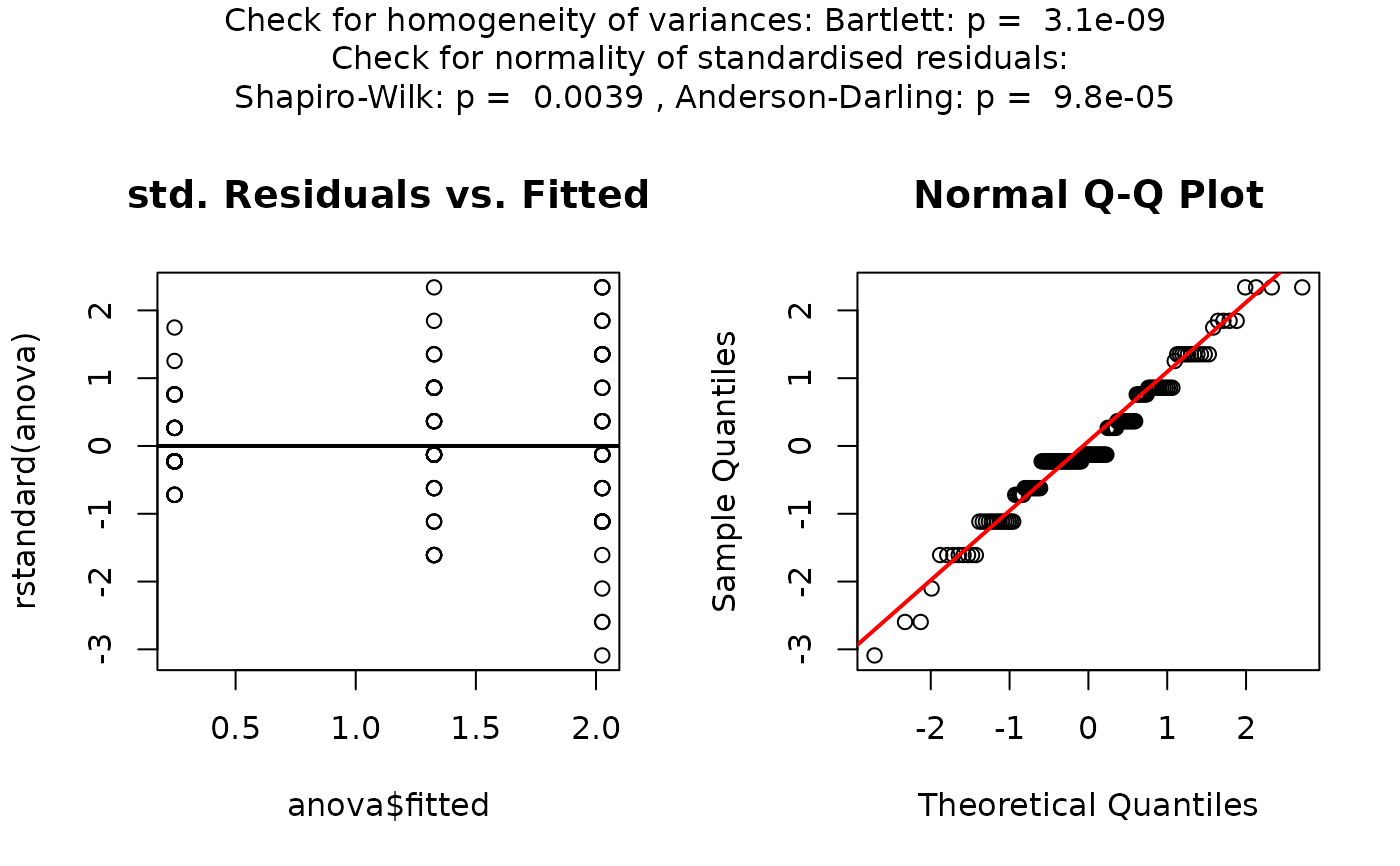
 ## Student's t-test (equal variances, two groups)
# When residuals are normally distributed and Levene's test indicates
# homoscedasticity, the classic Student's t-test with pooled variance is used
visstat(sleep$group, sleep$extra)
## Student's t-test (equal variances, two groups)
# When residuals are normally distributed and Levene's test indicates
# homoscedasticity, the classic Student's t-test with pooled variance is used
visstat(sleep$group, sleep$extra)

 ## Welch's t-test (unequal variances, two groups)
# When residuals are normally distributed but Levene's test indicates
# heteroscedasticity, Welch's t-test is used
visstat(mtcars$am, mtcars$mpg)
## Welch's t-test (unequal variances, two groups)
# When residuals are normally distributed but Levene's test indicates
# heteroscedasticity, Welch's t-test is used
visstat(mtcars$am, mtcars$mpg)
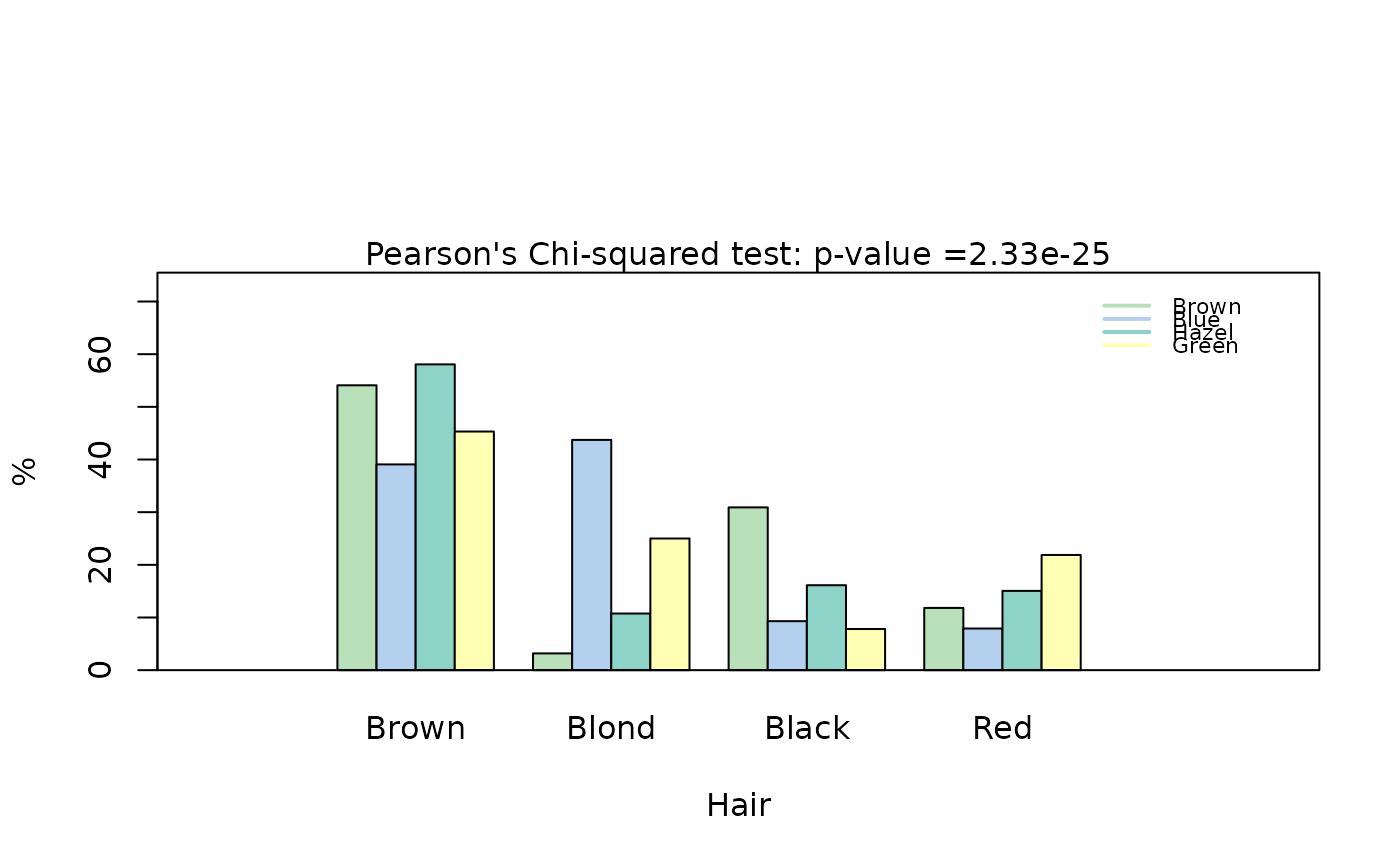
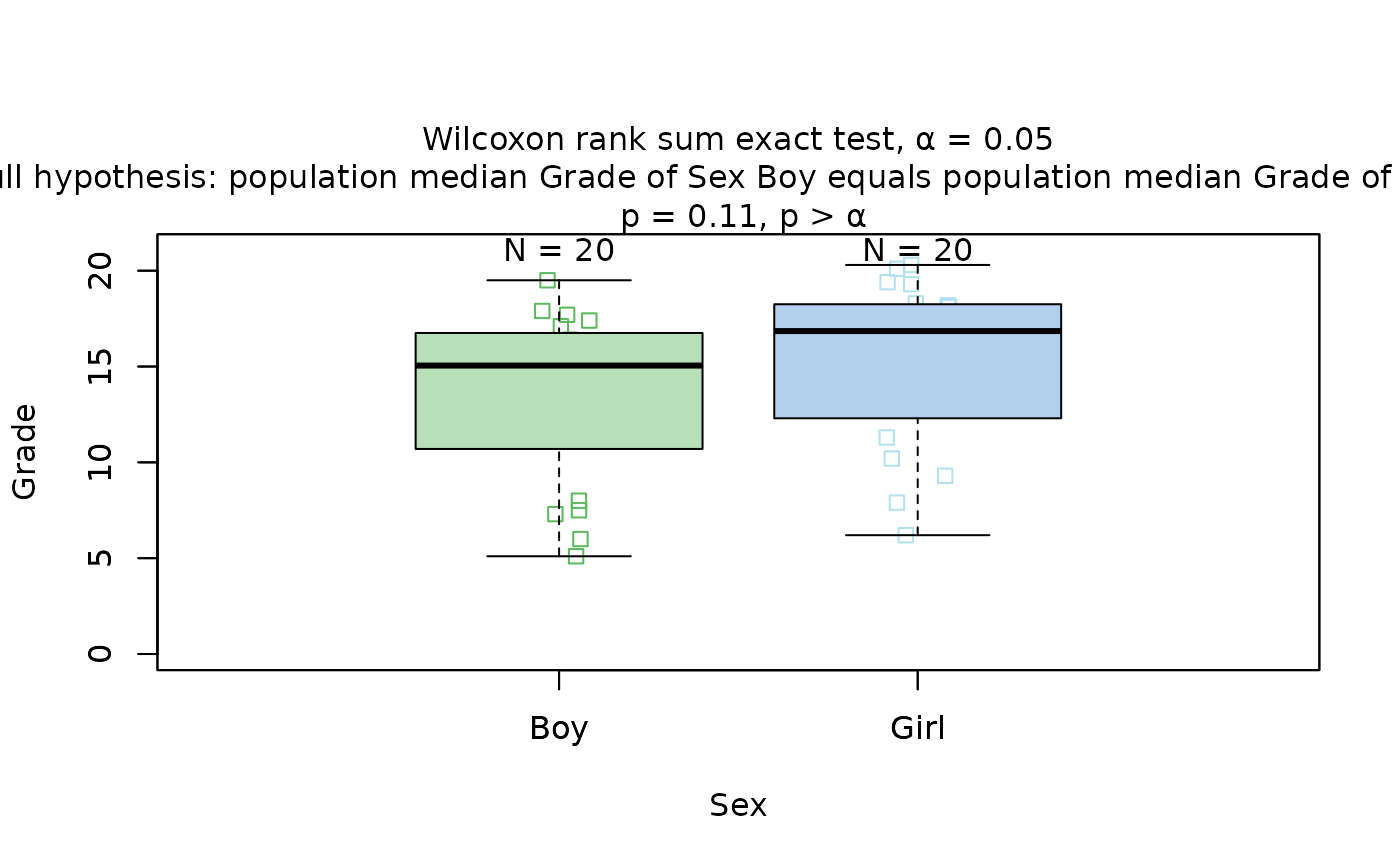
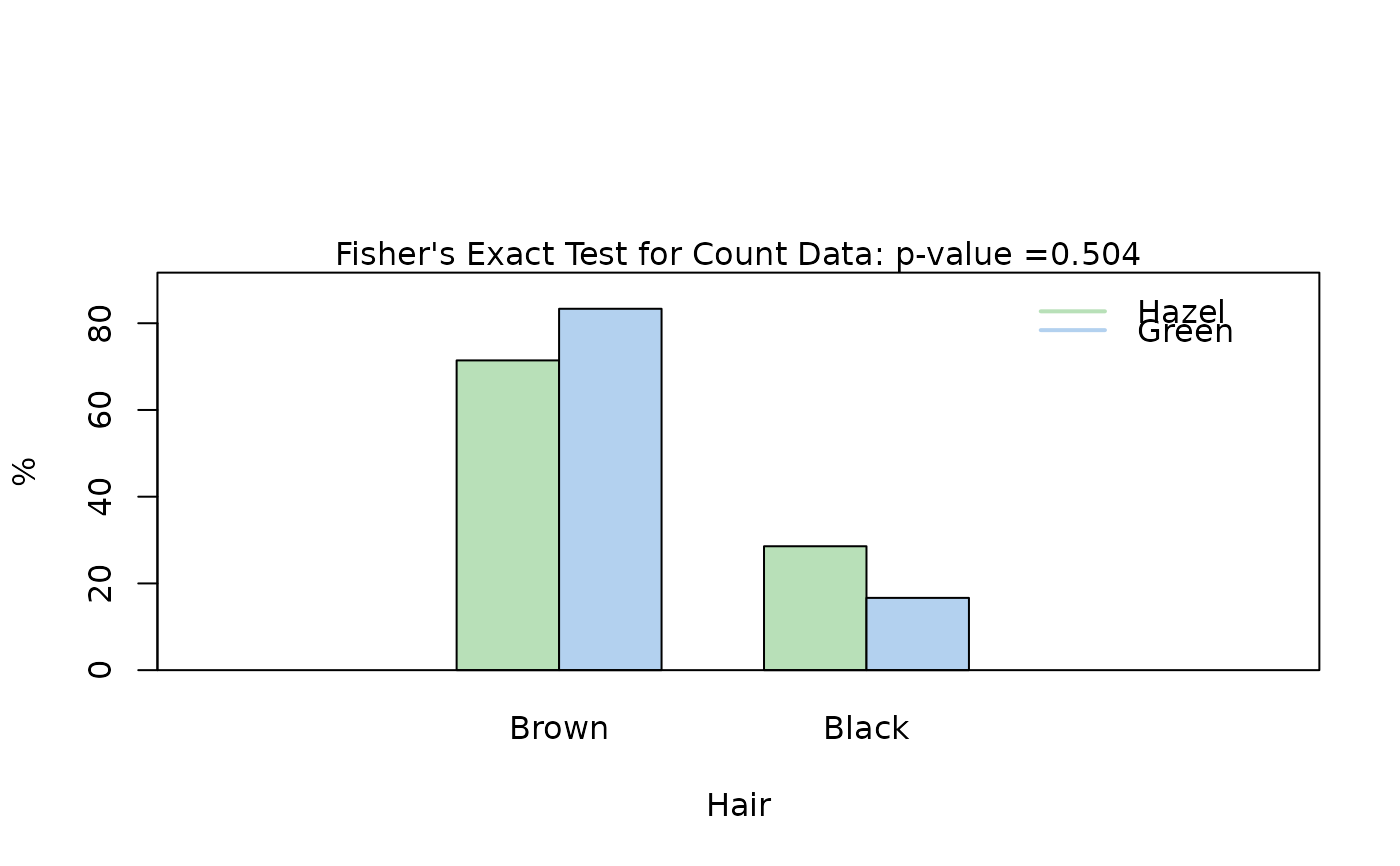 ## Wilcoxon rank sum test (non-normal, two groups)
# When residuals are not normally distributed
grades_gender <- data.frame(
Sex = as.factor(c(rep("Girl", 20), rep("Boy", 20))),
Grade = c(
19.3, 18.1, 15.2, 18.3, 7.9, 6.2, 19.4, 20.3, 9.3, 11.3,
18.2, 17.5, 10.2, 20.1, 13.3, 17.2, 15.1, 16.2, 17.3, 16.5,
5.1, 15.3, 17.1, 14.8, 15.4, 14.4, 7.5, 15.5, 6.0, 17.4,
7.3, 14.3, 13.5, 8.0, 19.5, 13.4, 17.9, 17.7, 16.4, 15.6
)
)
visstat(grades_gender$Sex, grades_gender$Grade)
## Wilcoxon rank sum test (non-normal, two groups)
# When residuals are not normally distributed
grades_gender <- data.frame(
Sex = as.factor(c(rep("Girl", 20), rep("Boy", 20))),
Grade = c(
19.3, 18.1, 15.2, 18.3, 7.9, 6.2, 19.4, 20.3, 9.3, 11.3,
18.2, 17.5, 10.2, 20.1, 13.3, 17.2, 15.1, 16.2, 17.3, 16.5,
5.1, 15.3, 17.1, 14.8, 15.4, 14.4, 7.5, 15.5, 6.0, 17.4,
7.3, 14.3, 13.5, 8.0, 19.5, 13.4, 17.9, 17.7, 16.4, 15.6
)
)
visstat(grades_gender$Sex, grades_gender$Grade)
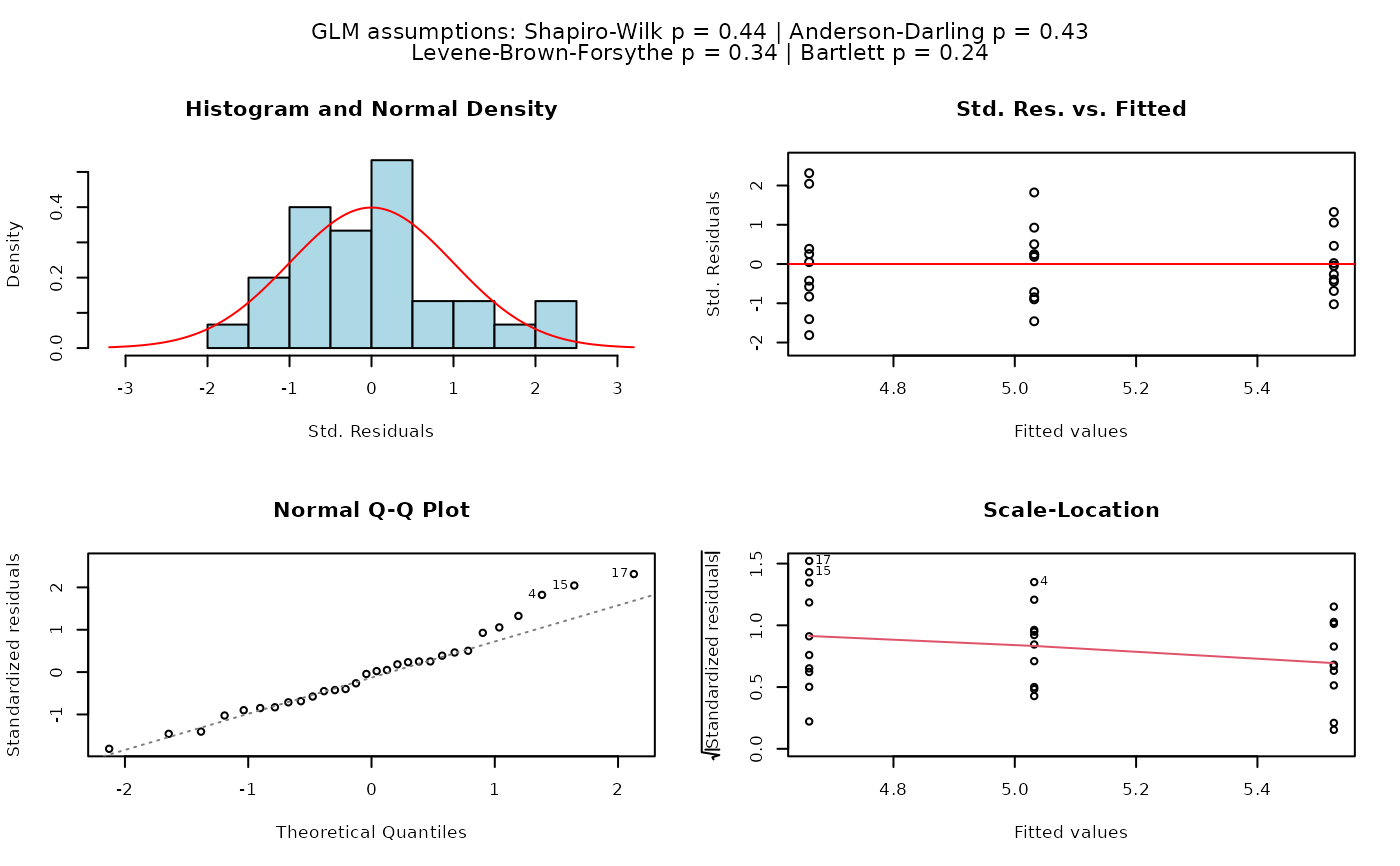
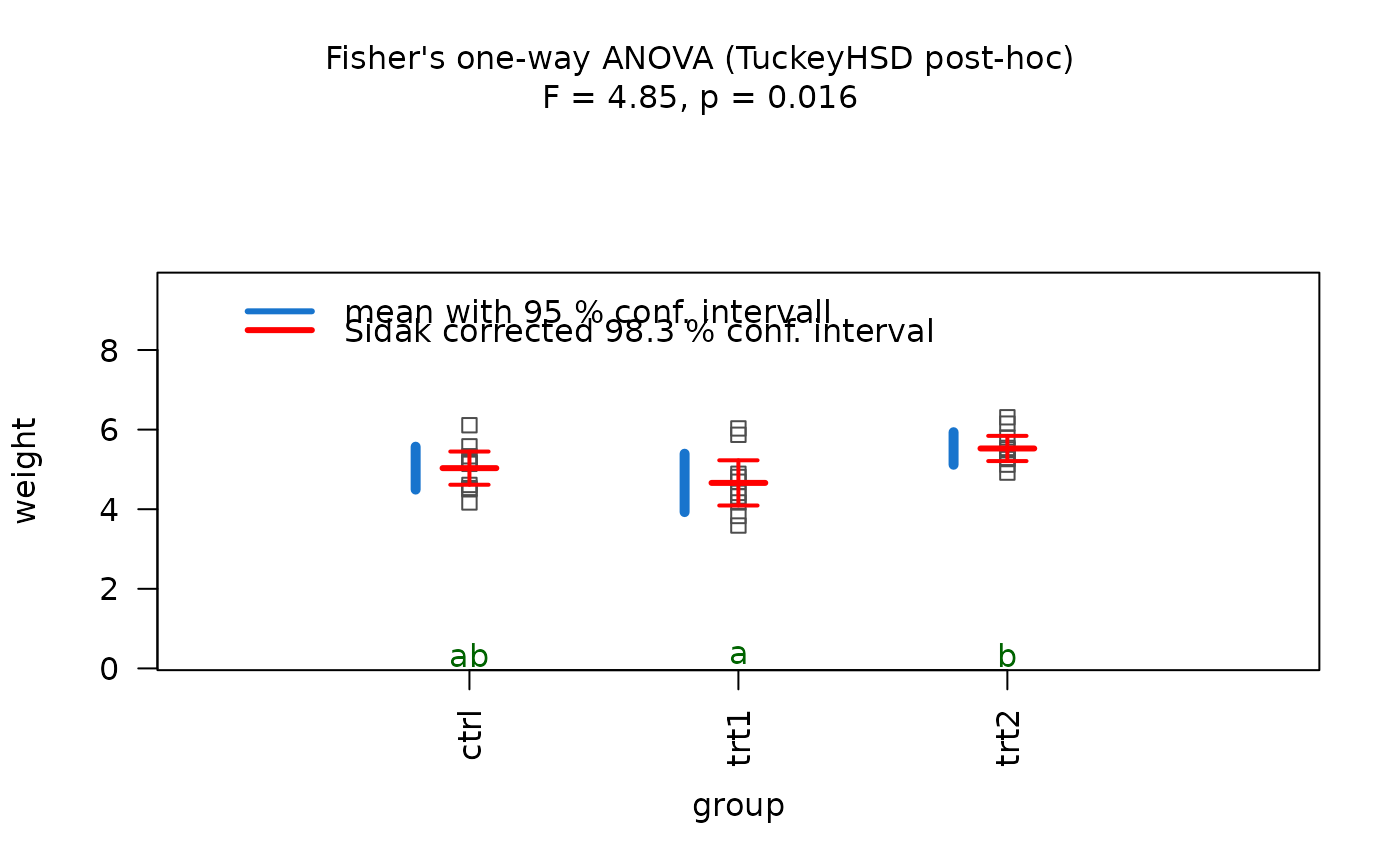
 ## Fisher's ANOVA (equal variances, >2 groups)
# When residuals are normally distributed and Levene's test indicates
# homoscedasticity, classic Fisher's ANOVA with TukeyHSD post-hoc is used
visstat(PlantGrowth$group, PlantGrowth$weight)
## Fisher's ANOVA (equal variances, >2 groups)
# When residuals are normally distributed and Levene's test indicates
# homoscedasticity, classic Fisher's ANOVA with TukeyHSD post-hoc is used
visstat(PlantGrowth$group, PlantGrowth$weight)
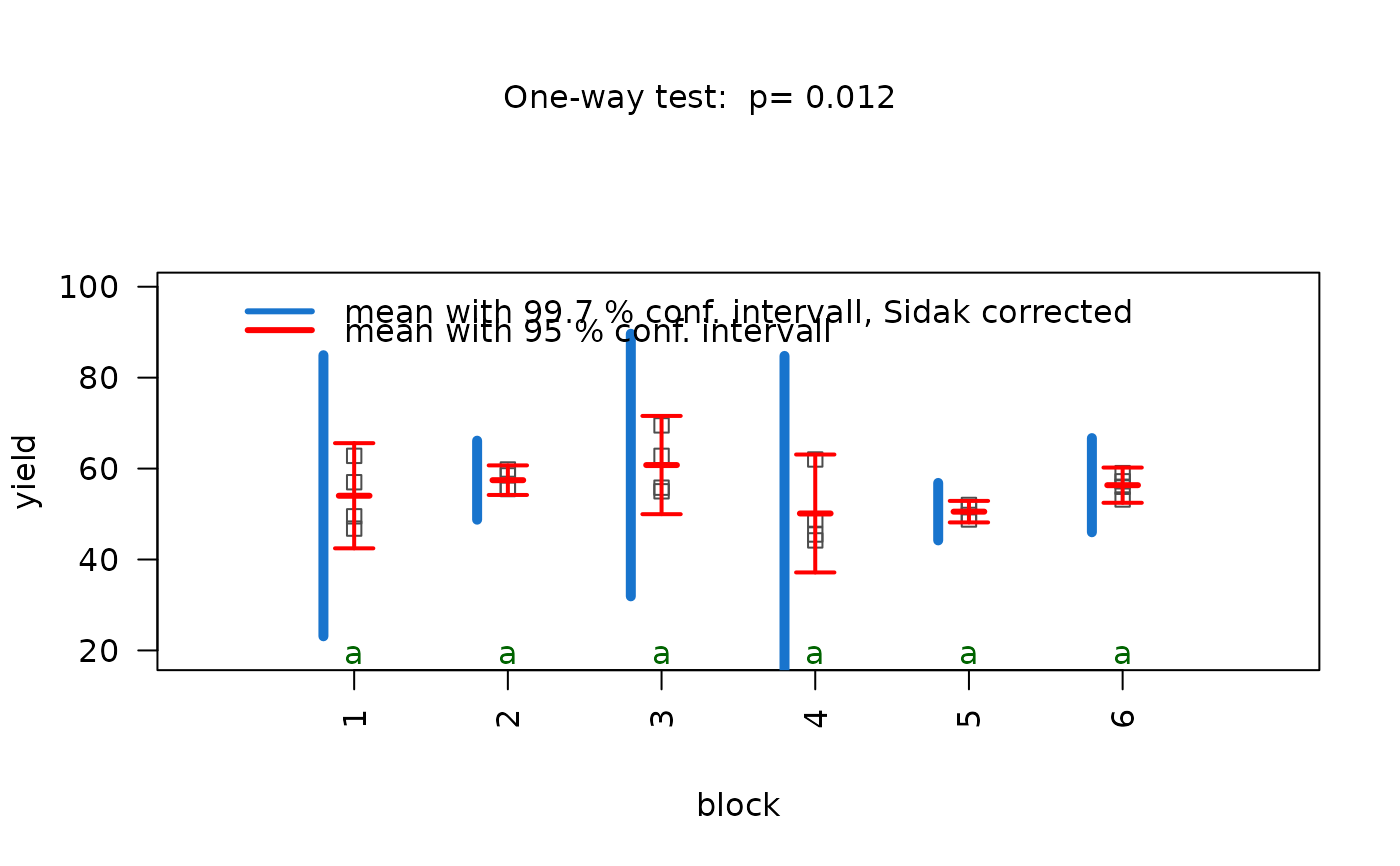
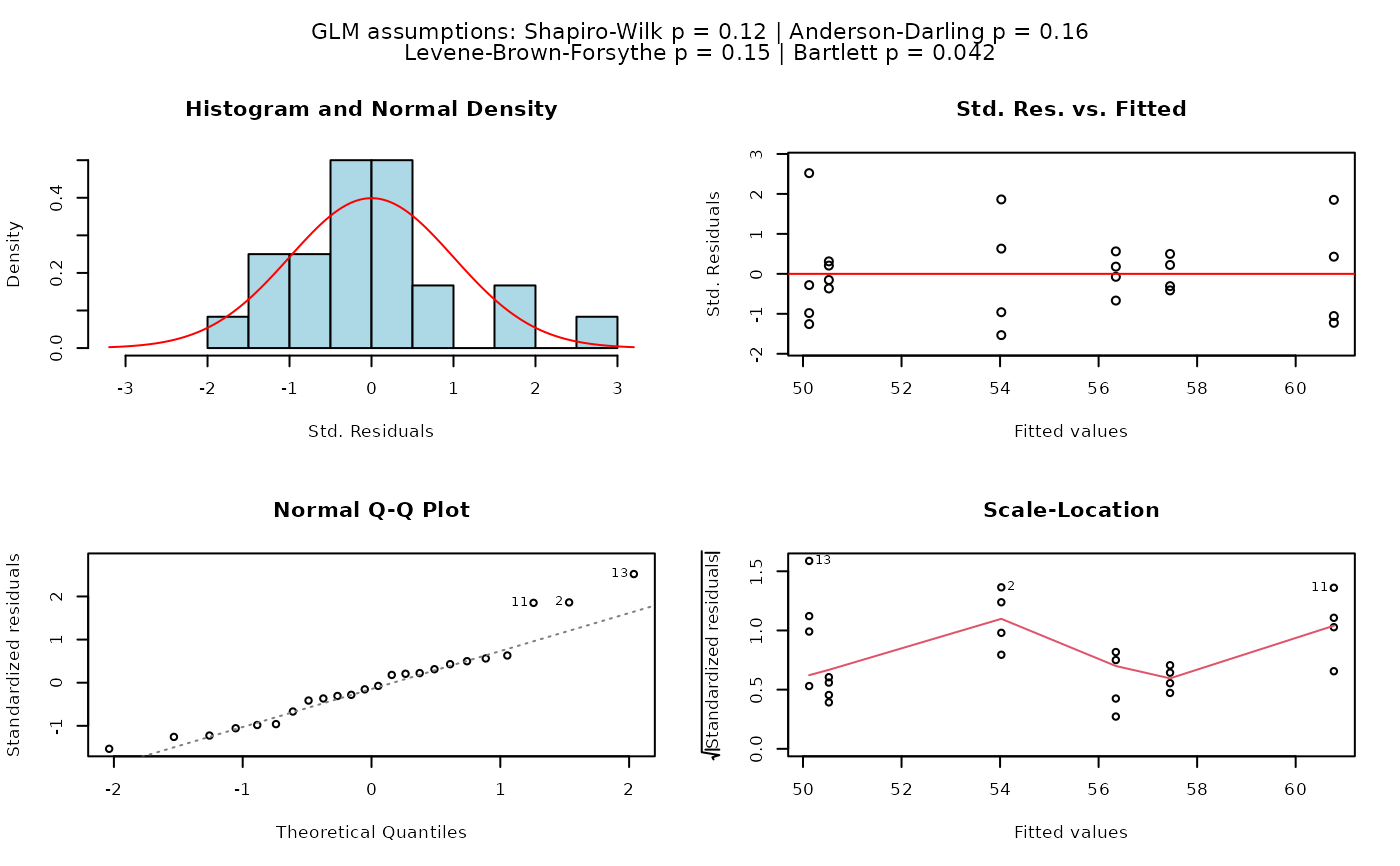
 ## Welch's one-way ANOVA (unequal variances, >2 groups)
# When residuals are normally distributed but Levene's test indicates
# heteroscedasticity, Welch's ANOVA with Games-Howell post-hoc is used
visstat(npk$block, npk$yield)
## Welch's one-way ANOVA (unequal variances, >2 groups)
# When residuals are normally distributed but Levene's test indicates
# heteroscedasticity, Welch's ANOVA with Games-Howell post-hoc is used
visstat(npk$block, npk$yield)
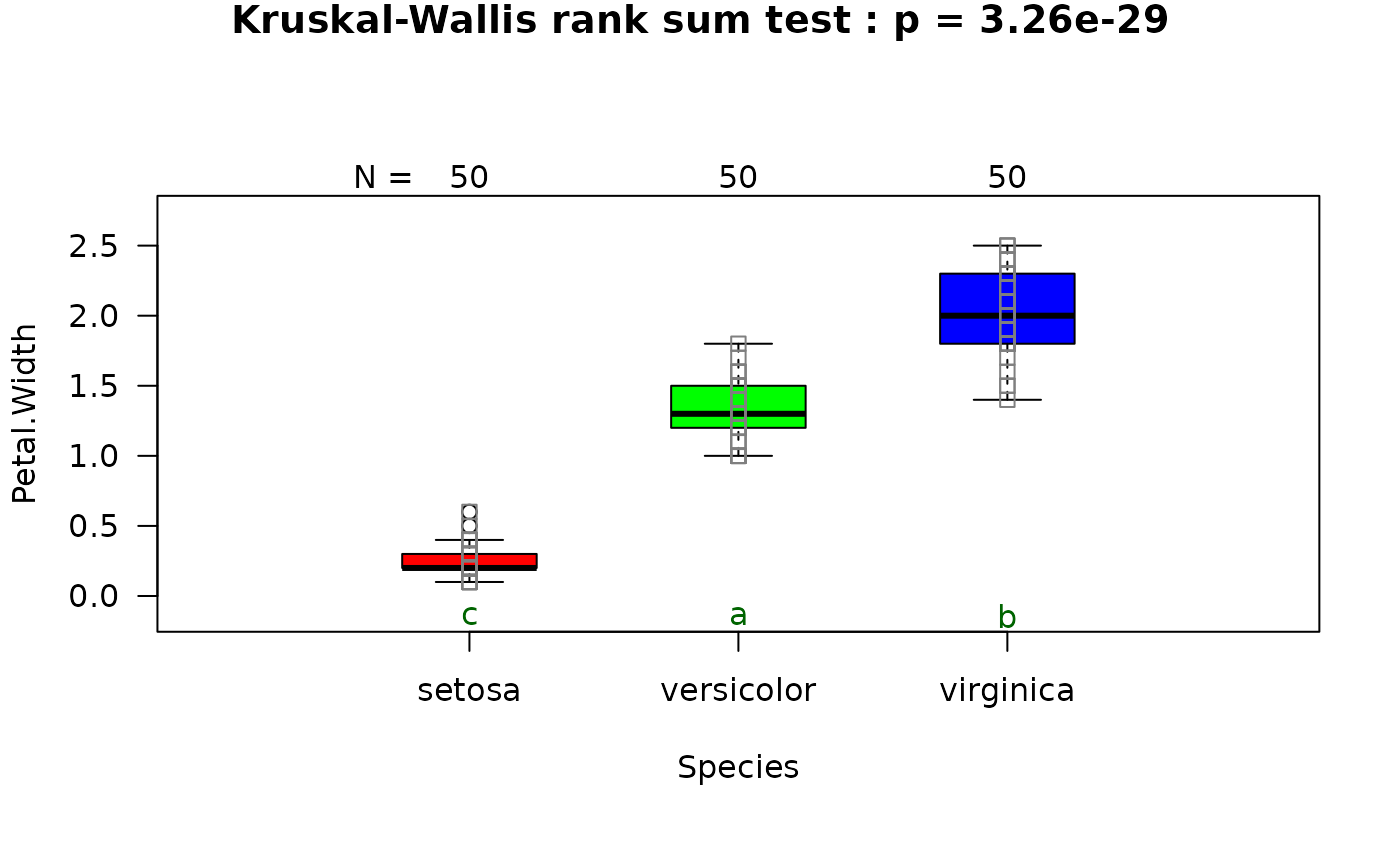
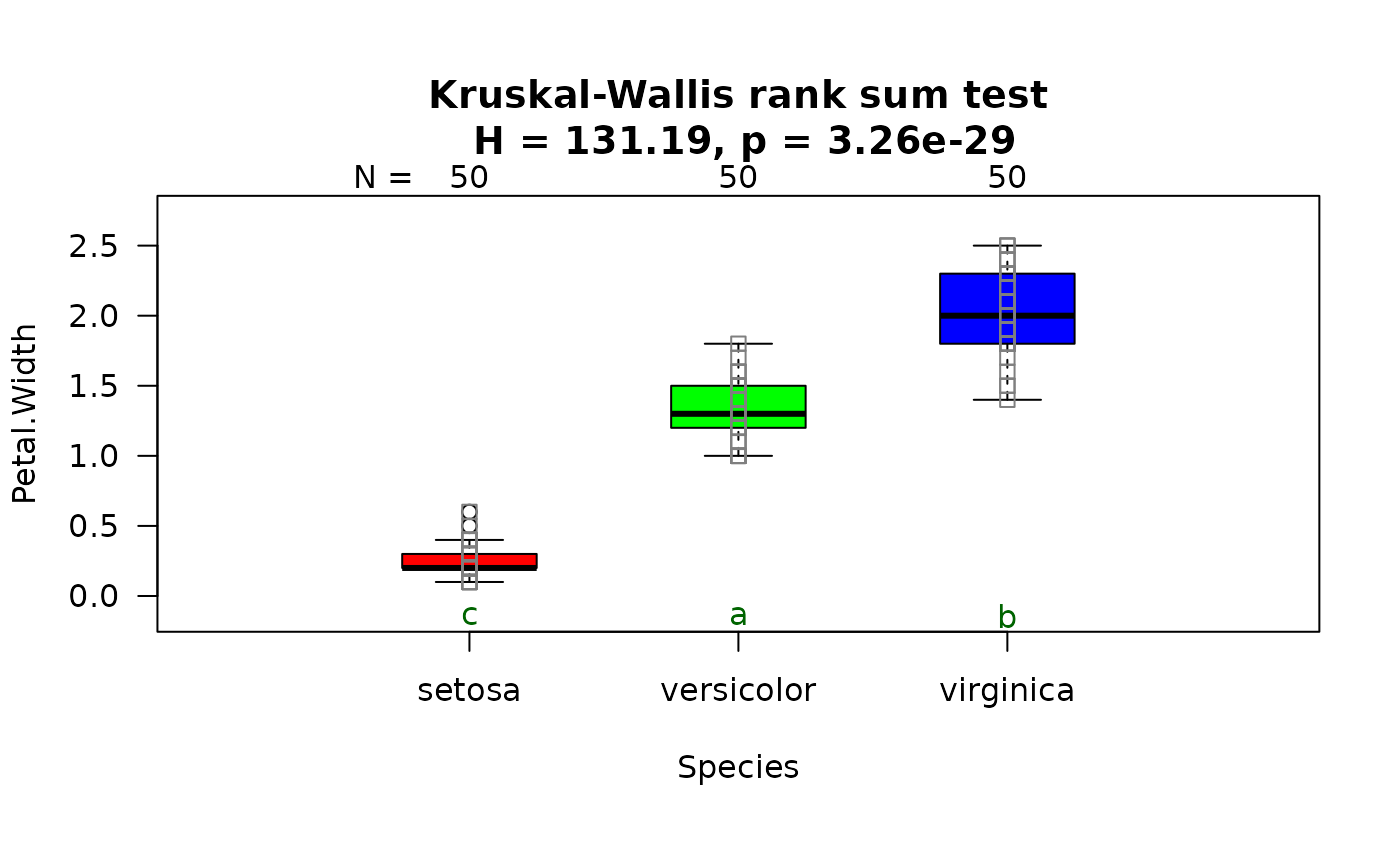
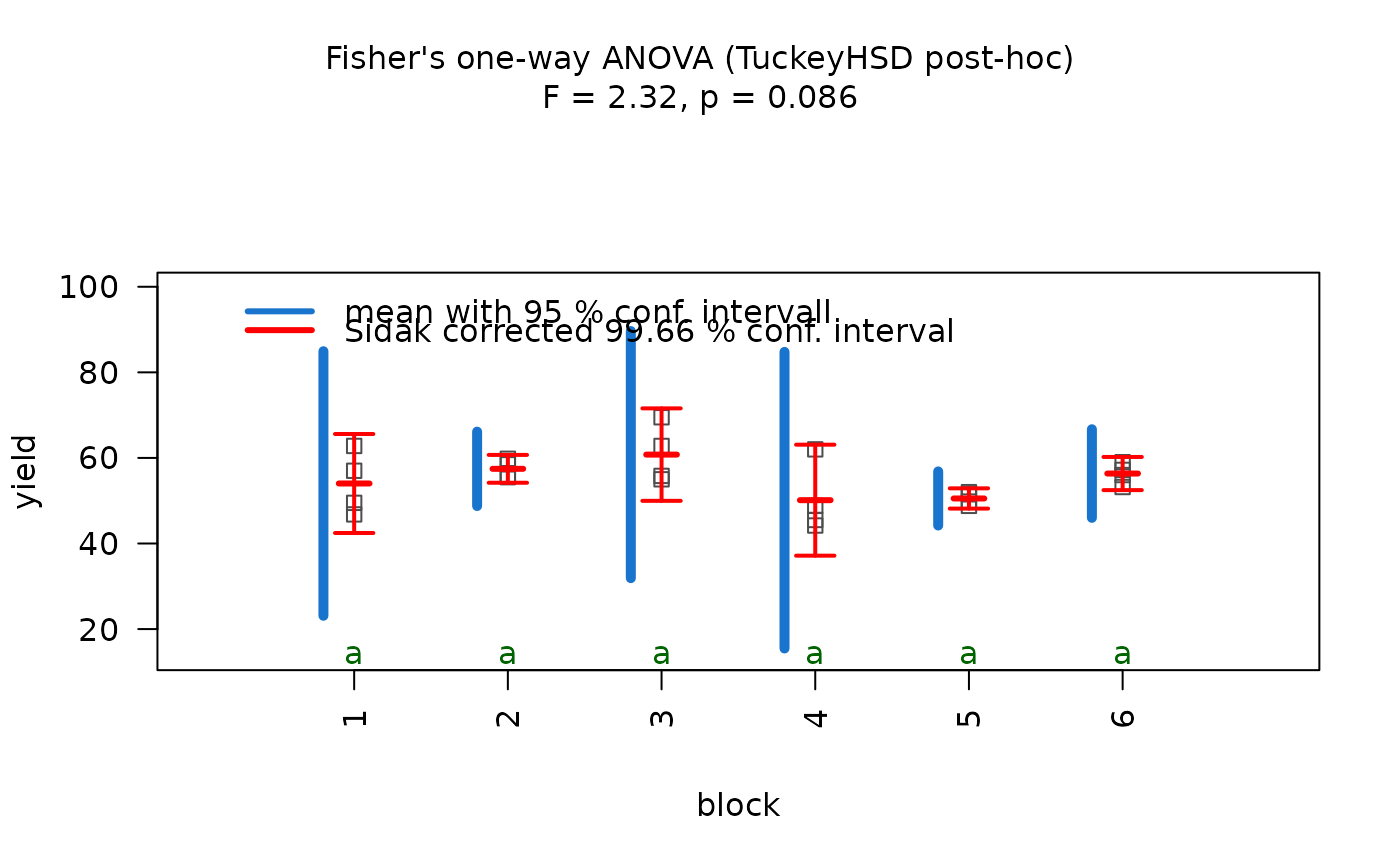 ## Kruskal-Wallis (non-normal, >2 groups)
# When residuals are not normally distributed
visstat(iris$Species, iris$Petal.Width)
## Kruskal-Wallis (non-normal, >2 groups)
# When residuals are not normally distributed
visstat(iris$Species, iris$Petal.Width)
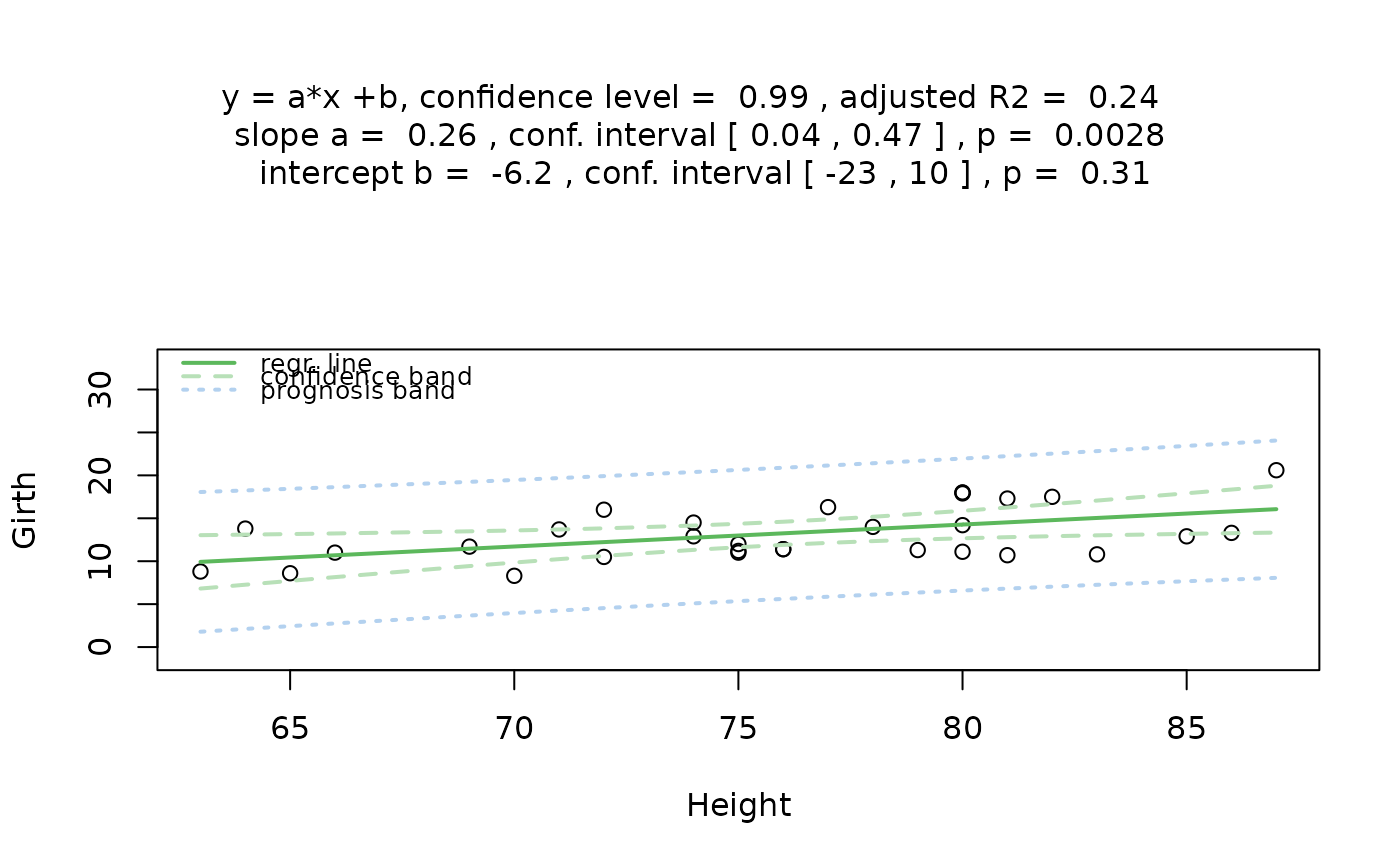
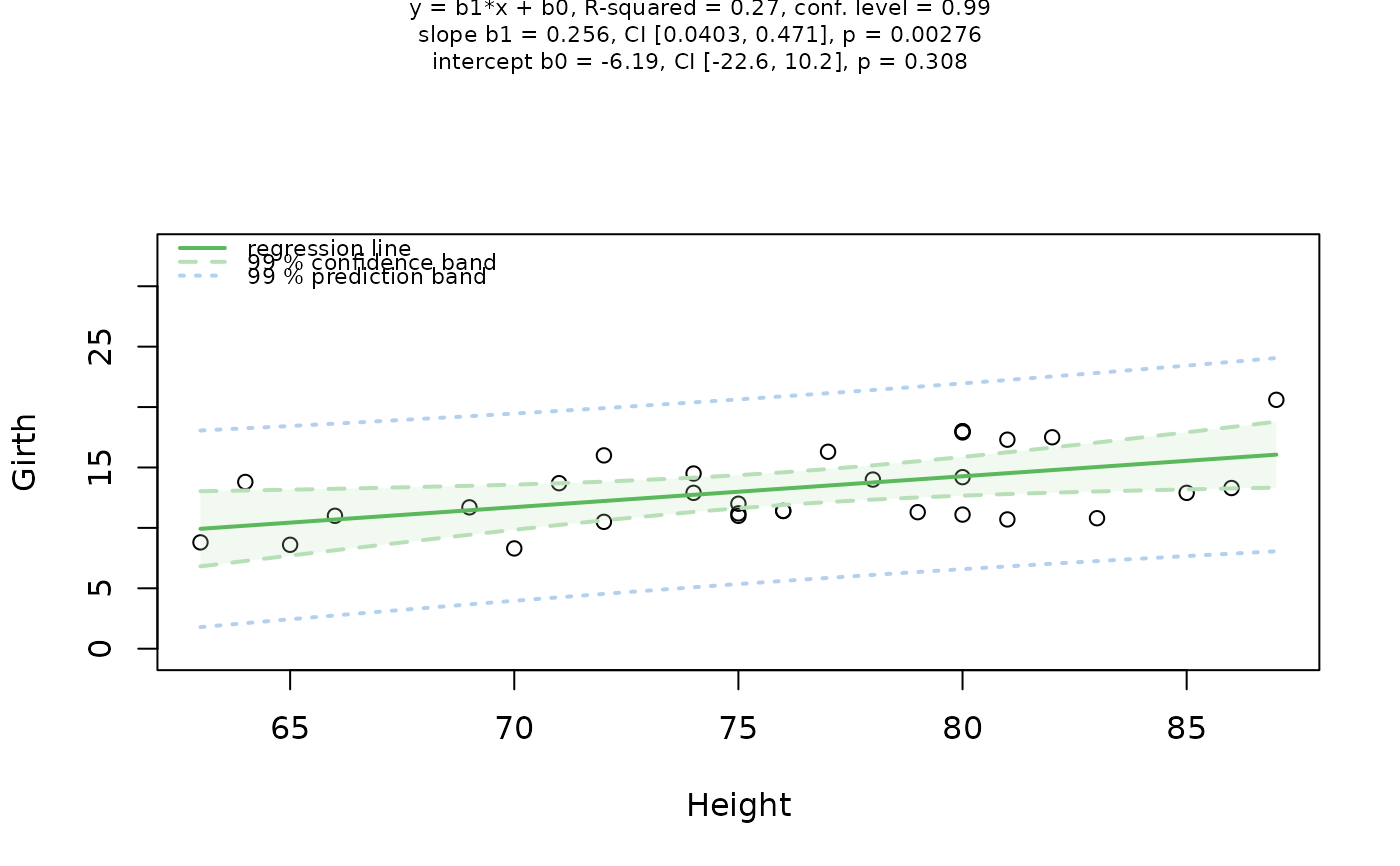
 ## Simple linear regression (both numeric)
visstat(trees$Height, trees$Girth, conf.level = 0.99)
## Simple linear regression (both numeric)
visstat(trees$Height, trees$Girth, conf.level = 0.99)
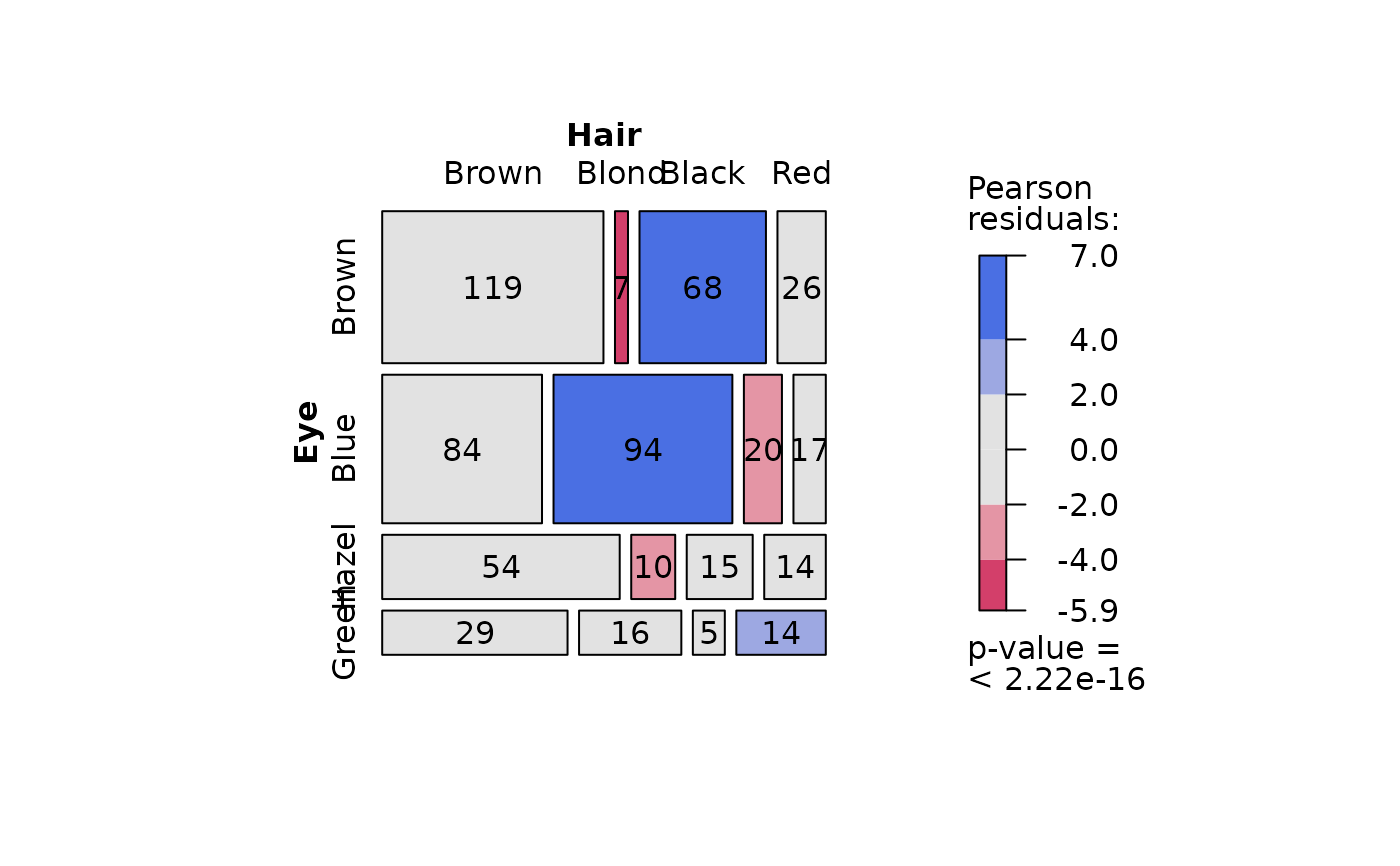
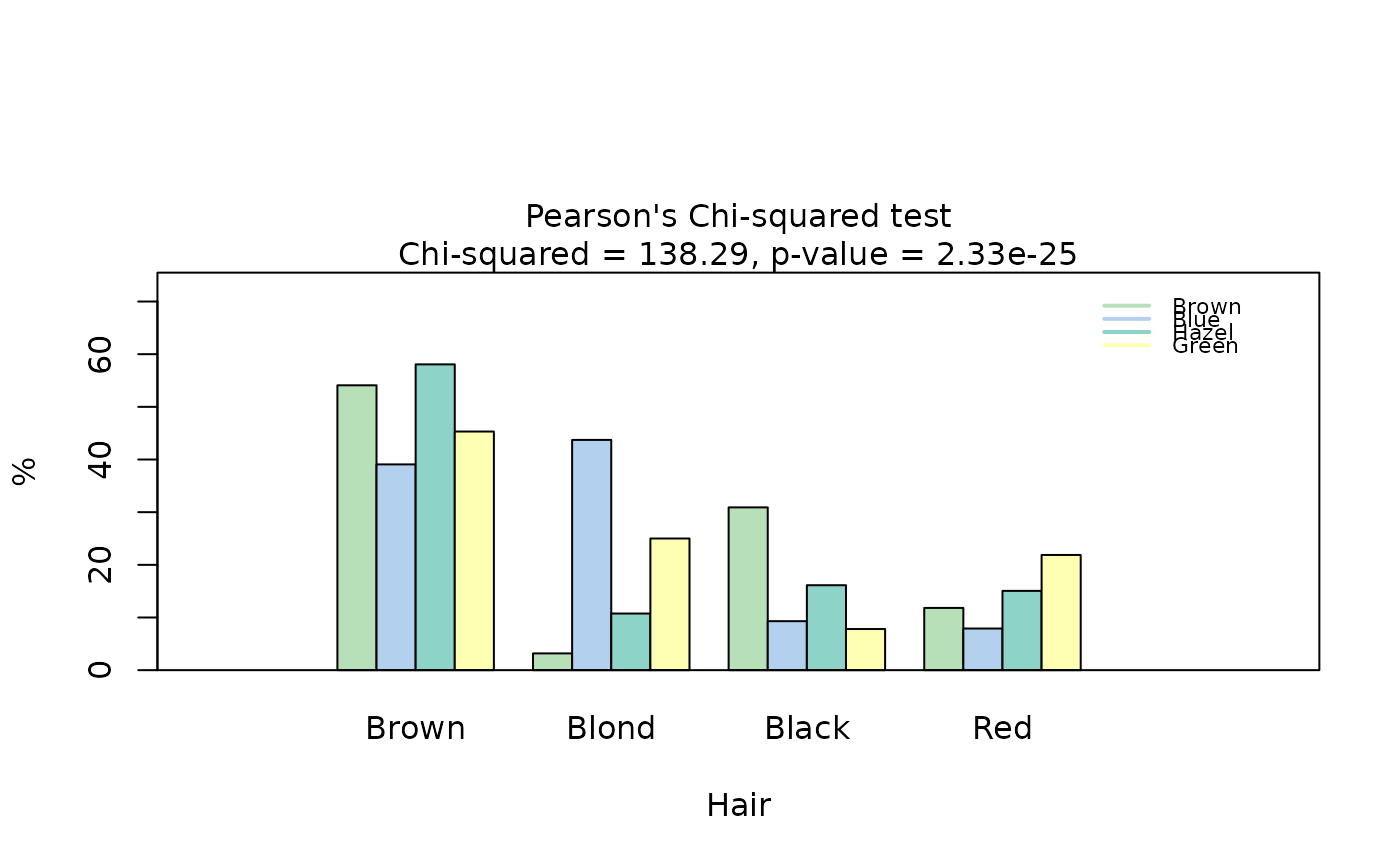
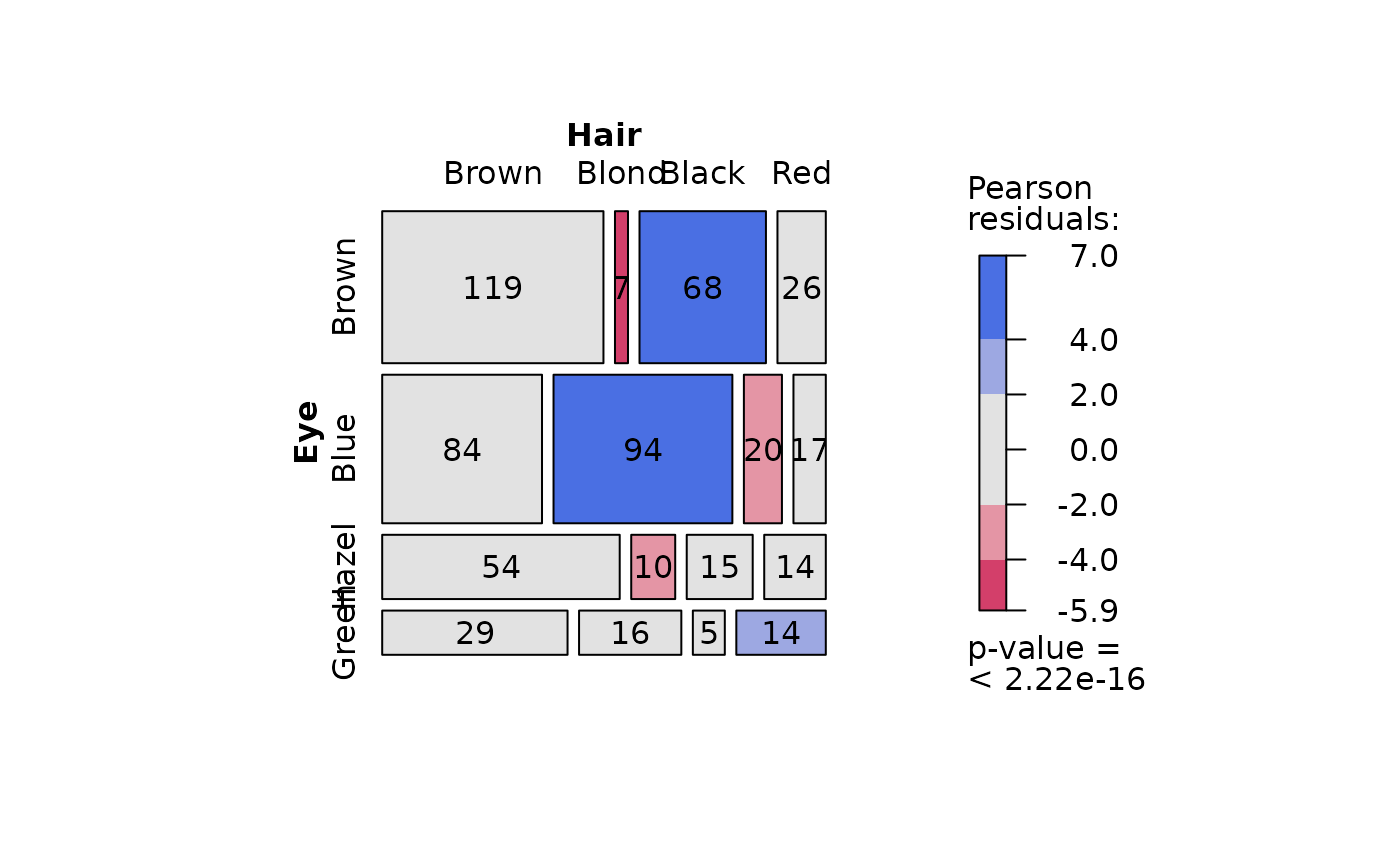
 ## Pearson's Chi-squared test (both factors, large expected counts)
HairEyeColorDataFrame <- counts_to_cases(as.data.frame(HairEyeColor))
visstat(HairEyeColorDataFrame$Eye, HairEyeColorDataFrame$Hair)
## Pearson's Chi-squared test (both factors, large expected counts)
HairEyeColorDataFrame <- counts_to_cases(as.data.frame(HairEyeColor))
visstat(HairEyeColorDataFrame$Eye, HairEyeColorDataFrame$Hair)
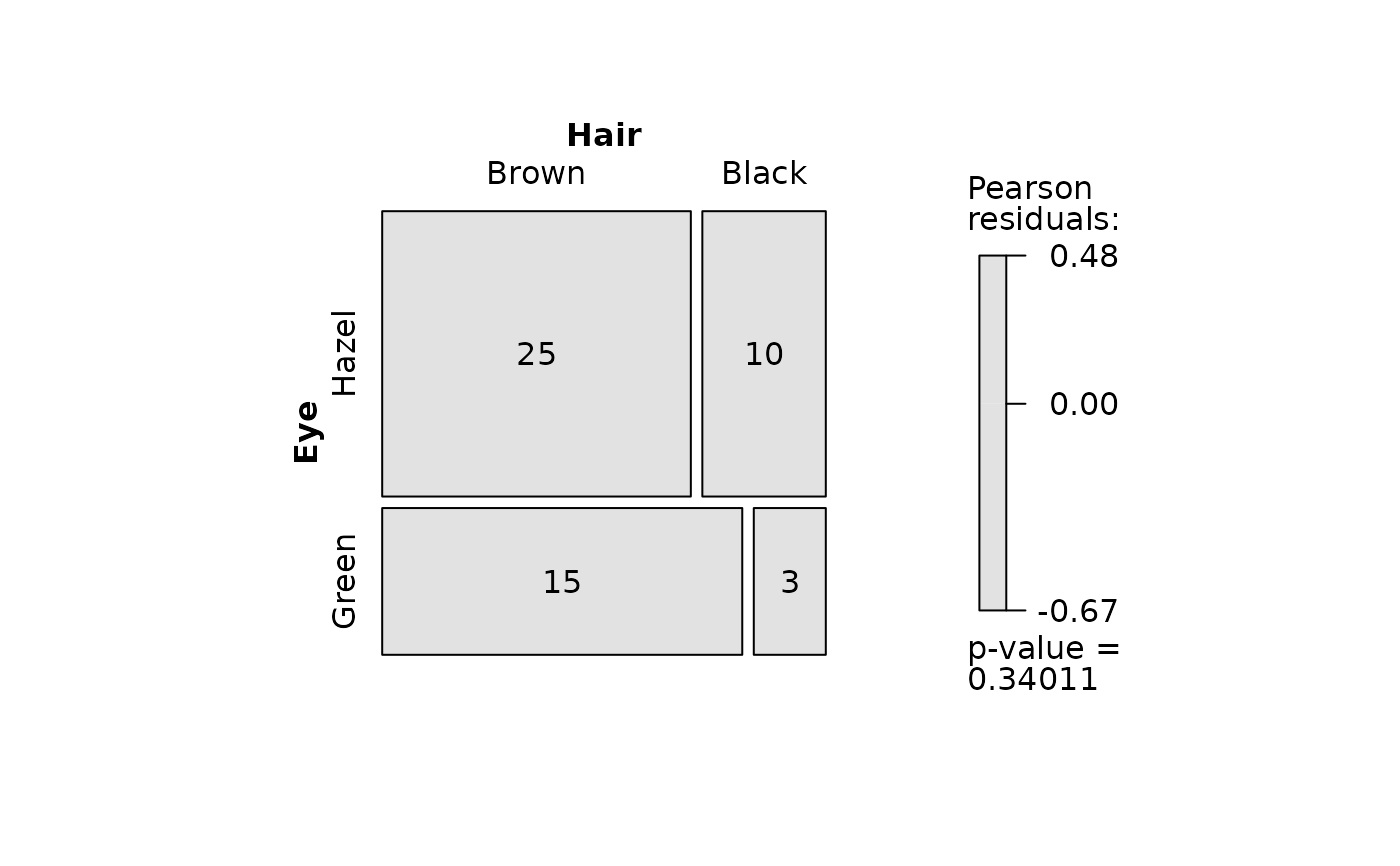
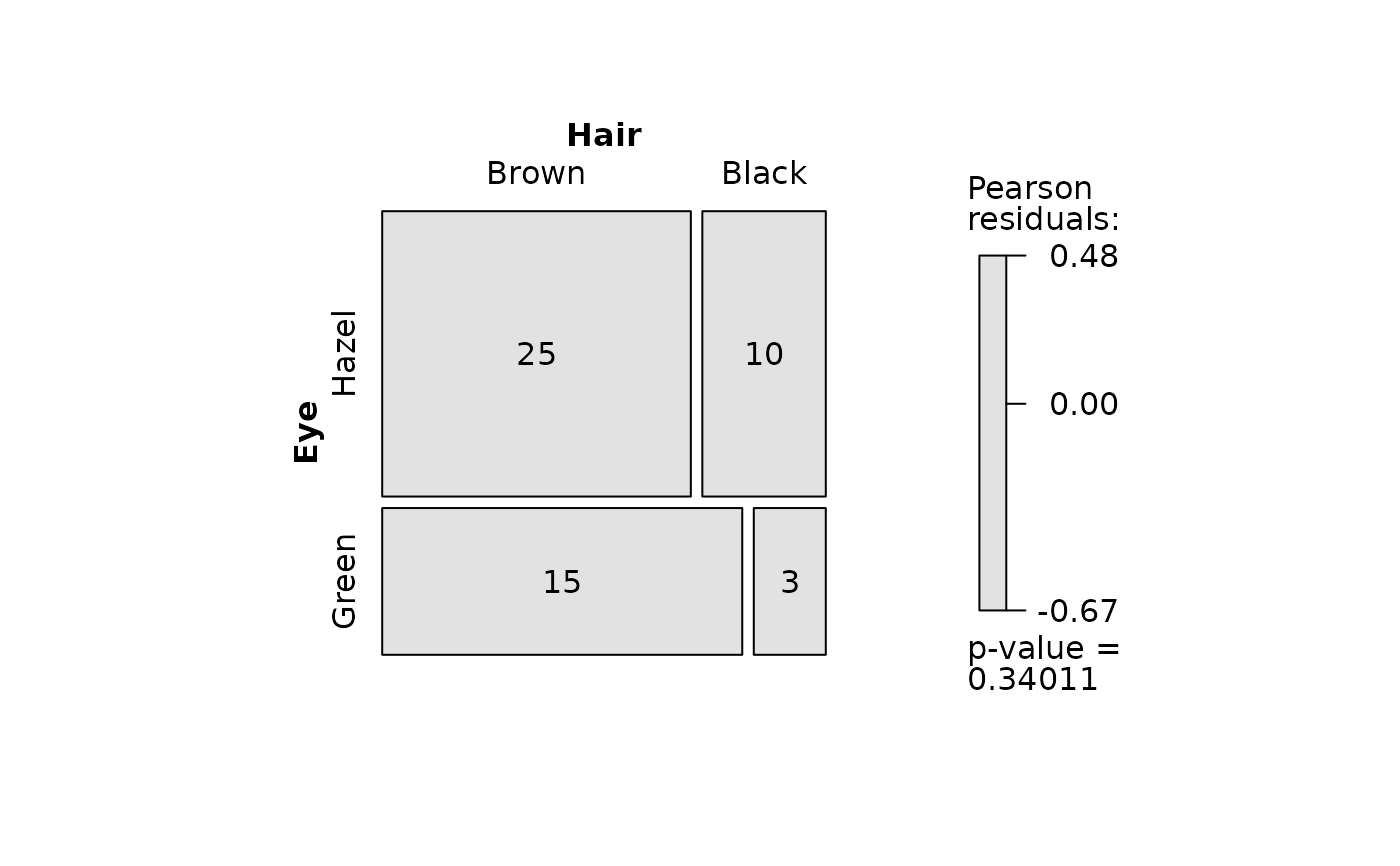
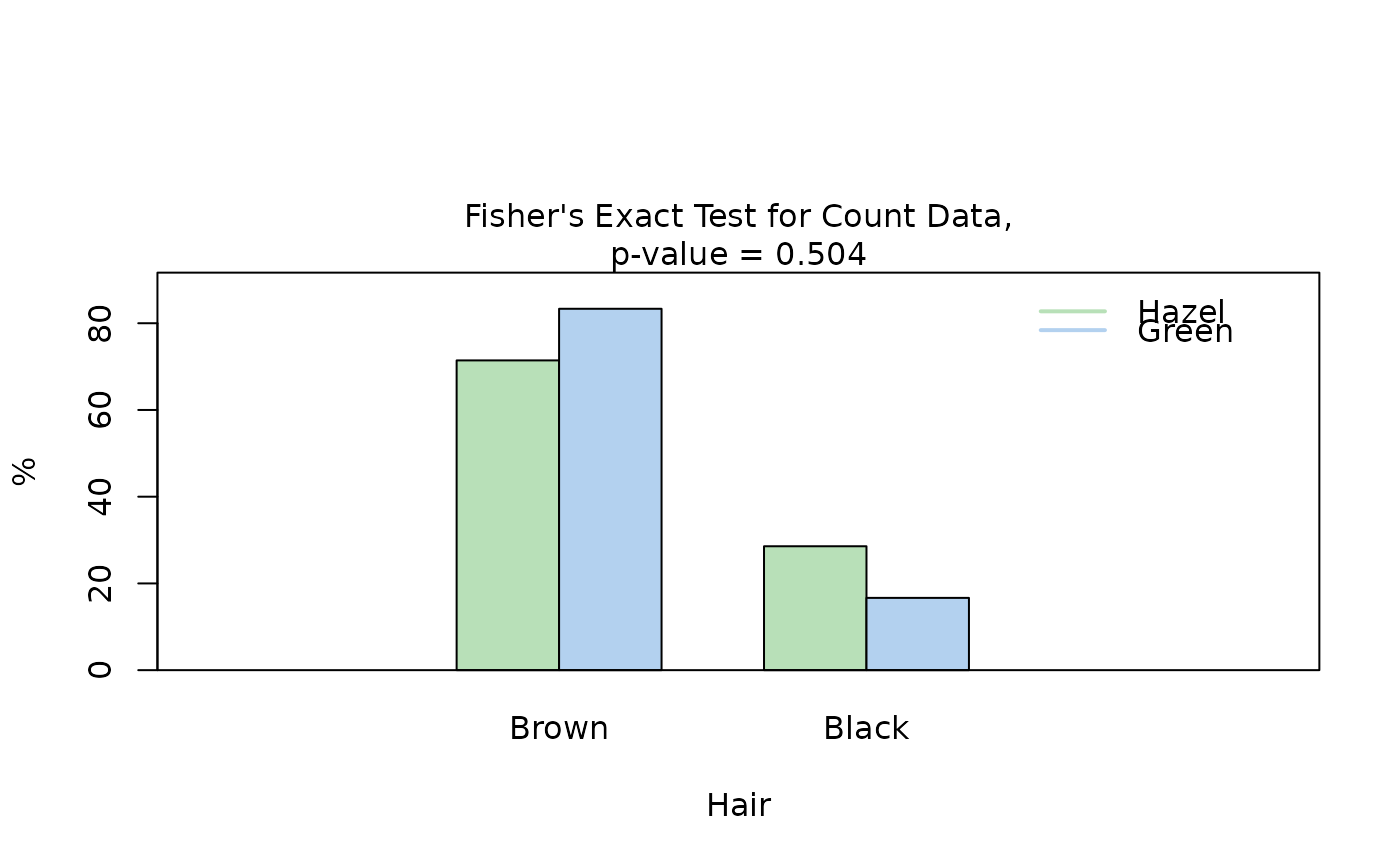
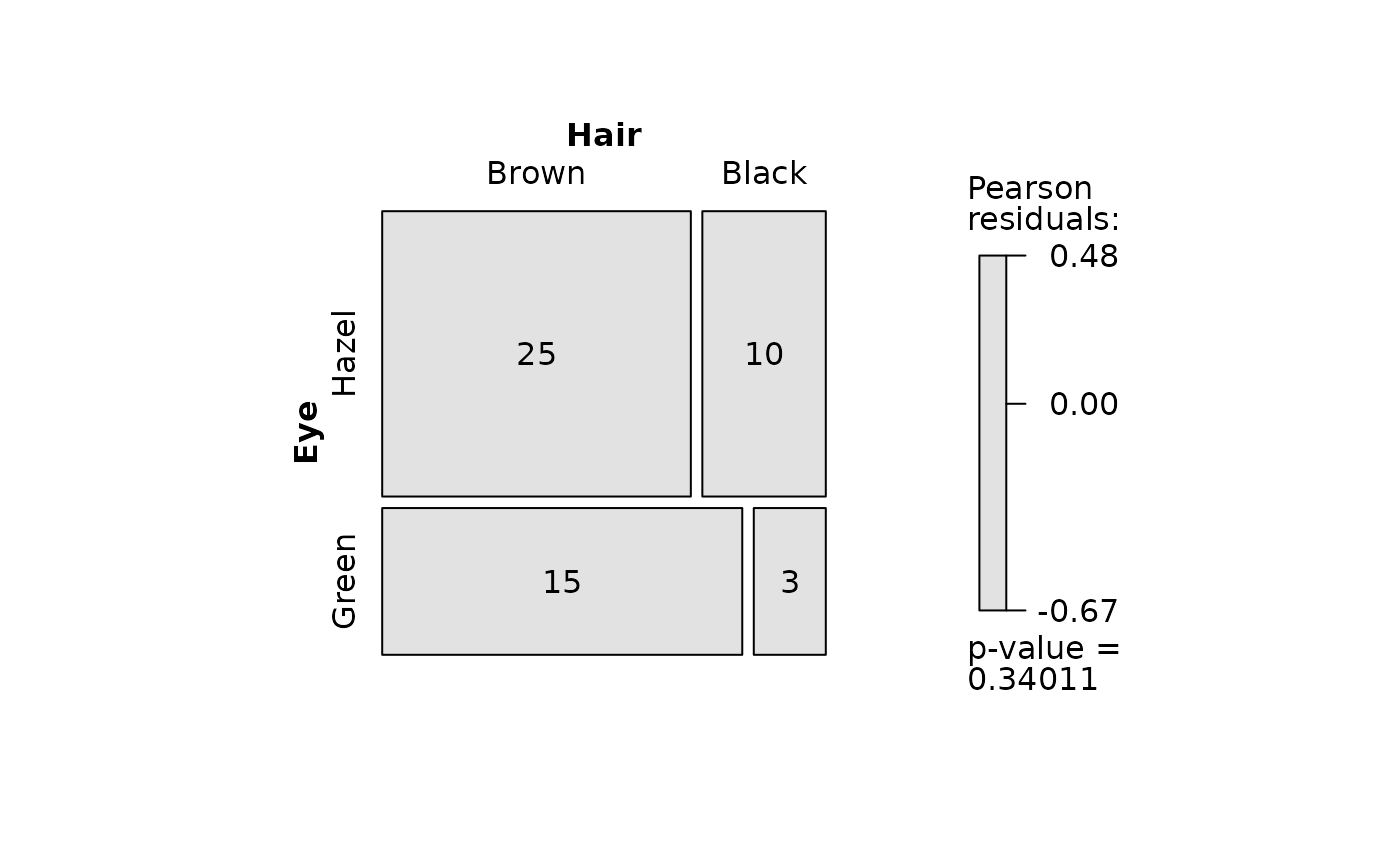
 ## Fisher's exact test (both factors, small expected counts)
HairEyeColorMaleFisher <- HairEyeColor[, , 1]
blackBrownHazelGreen <- HairEyeColorMaleFisher[1:2, 3:4]
blackBrownHazelGreen <- counts_to_cases(as.data.frame(blackBrownHazelGreen))
visstat(blackBrownHazelGreen$Eye, blackBrownHazelGreen$Hair)
## Fisher's exact test (both factors, small expected counts)
HairEyeColorMaleFisher <- HairEyeColor[, , 1]
blackBrownHazelGreen <- HairEyeColorMaleFisher[1:2, 3:4]
blackBrownHazelGreen <- counts_to_cases(as.data.frame(blackBrownHazelGreen))
visstat(blackBrownHazelGreen$Eye, blackBrownHazelGreen$Hair)
 ## Save PNG
visstat(blackBrownHazelGreen$Hair, blackBrownHazelGreen$Eye,
graphicsoutput = "png", plotDirectory = tempdir())
## Custom plot name
visstat(iris$Species, iris$Petal.Width,
graphicsoutput = "pdf", plotName = "kruskal_iris", plotDirectory = tempdir())
#> Warning: calling par(new=TRUE) with no plot
## Save PNG
visstat(blackBrownHazelGreen$Hair, blackBrownHazelGreen$Eye,
graphicsoutput = "png", plotDirectory = tempdir())
## Custom plot name
visstat(iris$Species, iris$Petal.Width,
graphicsoutput = "pdf", plotName = "kruskal_iris", plotDirectory = tempdir())
#> Warning: calling par(new=TRUE) with no plot